
Introduction
The German cDNA Consortium (1) within SMP-Cell systematically identifies and clones novel genes and their protein coding regions, and thus continues to contribute significantly to the international effort to generate and provide clone resources of human genes, splice forms and protein coding regions to the scientific community. The major focus of the consortium is on disease relevant genes and ORFs that are identified in collaborating projects within KGs and SMPs.
 | |
| Fig 1: HTS-Assays cover cell cycle progression and apoptosis. | |
The clone resources thus serve the immediate exploitation in the functional genomics pipeline within SMP-Cell (2), and for collaborating SMPs (e.g. SMP-RNA and SMP-Protein), and KGs.
Currently, only ~ 60% of the estimated 24,000 human genes are available as full-length cDNAs to the scientific community. In many cases sequences and clones, although being annotated full-length, are truncated, and miss significant parts especially at their N-terminal ends. Moreover, many genes have not been cloned at all.
Project Status
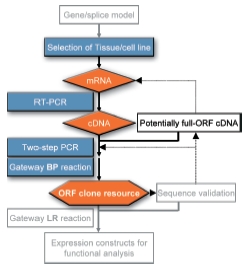
As the novelty rate in random sampling of cDNA libraries has dramatically decreased over the years, the strategy of the cDNA consortium was changed from the sampling of such libraries to the systematic modeling and cloning of full length genes and open reading frames. The selected target genes are mostly long and rarely expressed genes with disease relevance. While gene modeling is carried out only at the DKFZ, the cloning process (Fig. 1) is performed in several locations; AGOWA GmbH Berlin, DKFZ Heidelberg, Qiagen GmbH Hilden, RZPD Heidelberg, and the BMFZ of the University Düsseldorf. This distribution of cloning increases the throughput that is possible, while a strict quality control and standardization of protocols has been established to guarantee high quality standards of the final clone resource.
 | |
| Fig 2: Assembly of processes in the generation of ORF resources: A) Interface of ORF cloning database, B) comparison of PCR products of first and second step ORF amplification, C) analysis of entry clone sequences, D) example of cloning step automation. | |
ORFs and splice variants are primarily amplified via RT-PCR from RNA of either cell lines or tissues that have been identified to express the desired ORF/splice variant. In addition, the cDNA clones that have been EST-sequenced in large scale projects, including that of the German cDNA Consortium in DHGP and NGFN-1, is a rich source for potential full-ORF cDNAs that are utilized whenever possible as templates for amplification (Fig. 1). PCR products (Fig. 2) are cloned into the Gateway® system (Fig. 1) and the resulting entry clones are sequence verified for further processing in the functional analysis pipeline (3).
An ORF resource of more than 1,200 sequence-verified Gateway entry clones was generated within DHGP and NGFN-1, having an average ORF size of > 2 kb. The ORF amplification and cloning procedure has been continuously optimized and includes a high level of automation (Fig. 2). Different DNA polymerases have been tested for their fidelity and processivity in view of low error rates in ORF amplification. SOPs have been established and are employed to achieve and maintain high throughput and quality standards in the ORF-clone resources. Data management and quality control of the cloning process is guaranteed by the implementation of a LIMS database (4) (Fig. 2). It is designed to automatically generate and maintain a standardized nomenclature during all steps of the cloning process, and is essential to allow for the tracking of expression constructs (and the functional data generated with them later) to materials and sequences. The software generates PCR IDs, entry clone IDs, expression clone IDs, and automatically assembles 96-well plates for sequence validation and for further processing (Fig. 2). The complete sequence validation of the ORF-inserts and the annotation of any deviations from the presumed consensus sequence have proven to be essential in the quality control process. Potential frame-shift mutations which could be introduced during the ORF amplification process need to be identified and necessitate re-cloning of the respective ORF. The cloning process is therefore tightly connected to the sequence validation that is carried out in another project of SMP-Cell (Fig. 1). Finally, only validated clones are entered into the functional analysis pipeline of the cell-based assays of SMP-Cell and of collaborating SMPs and KGs.
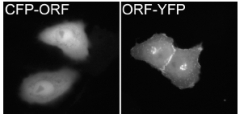 | |
| Fig 3: Example of protein localizing to Golgi apparatus and plasma membrane. The orientation of the GFP-tag relative to the ORF impacts the localization of fusion proteins. Protein CFP-ORF localizes artificially due to the color tag at its N-terminus, while the YFP-tag does not interfere with the proteinís localization when expressed at the C-terminal end. | |
In this line, all ORFs were initially amplified and cloned lacking the original stop codon in the PCR product during DHGP and NGFN-1. This enabled the expression of N- and C-terminal fusion proteins from one entry clone. However, the lack of the native stop codon introduced a few additional residues that were artificially added at the C-terminus of the encoded proteins. We have changed our strategy in NGFN-2 to now clone two entry clones for every ORF, one with and the other without the native stop codon. N-terminally tagged proteins and natively expressed proteins in consequence terminate at their natural C-terminus. To this end the first PCR in the ORF amplification process is performed with primers that are degenerate in one position, one species containing and the other lacking a stop codon. The selection for either species is done in the second PCR step, where two separate reactions are performed for either product. The amplification and cloning procedures have been standardized, the SOPs are freely available at the SMP-Cell web site at http://www.smp-cell.org.
Outlook
The generation of comprehensive ORF clone collections has become a key challenge in functional genomics and proteomics. With the changed strategy from random cDNA library sampling to the modeling and cloning of defined genes, the ORF clone resources of the German cDNA Consortium within SMP-Cell are efficiently extended in NGFN-2 and provide the basis for functional exploitation of the encoded proteins within SMP-Cell and in collaborating SMPs and KGs. The amplification and cloning of the ORFs will be continuously optimized to further reduce error rates, especially for the RT-PCR cloning of ORFs that are not covered by any cDNAs yet. This guarantees the production of a standardized and quality-controlled resource for the successful screening of the encoded proteins in the disease-relevant cell-based assays within the SMP-cell (5, 6), and by scientists in collaborating SMPs and KGs.
Lit.: 1. Wiemann S et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001 Mar;11(3):422-35. 2. Wiemann S et al. From ORFeome to biology: a functional genomics pipeline. Genome Res. 2004 Oct;14(10B):2136-44. 3. Wiemann S et al. The German cDNA network: cDNAs, functional genomics and proteomics. J Struct Funct Genomics. 2003 4(2-3):87-96. 4. Bannasch D et al. LIFEdb: a database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system. Nucleic Acids Res. 2004 Jan 1;32:D505-8. 5. Arlt D et al. Functional profiling: from microarrays via cell-based assays to novel tumor relevant modulators of the cell cycle. Cancer Res. 2005 in press. 6. Starkuviene V et al. High-content screening microscopy identifies novel proteins with a putative role in secretory membrane traffic. Genome Res. 2004 Oct;14(10A):1948-56.


