
Introduction
cDNAs are indispensable tools for the identification of genes, even with several genome sequences available today. They are key to defining splice variants and form the basis for functional gene analysis. The German cDNA Consortium was second worldwide to carry out the systematic full-length sequencing of human cDNAs. This consortium is a highly successful example of a large scale and long time collaboration of academic and commercial institutes (1-4):
Sequence validation of ORF clones is performed at AGOWA (Dagmar Heubner), DKFZ (Stefan Wiemann), GBF (Helmut Blöcker), Medigenomix (Birgit Ottenwälder), Qiagen (André Bahr) and the University of Düsseldorf (Karl Köhrer).
 | |
| Fig 1: The H-invitational annotation jamboree was organized in Tokyo. Scientists from 60 institutions worldwide manually annotated full-length cDNAs from the large-scale cDNA projects. The German cDNA Consortium was represented with sequences and scientists (11). (photo: Dr. Takashi Imanishi) | |
Project Status
The general strategy of the German cDNA Consortium has been changed in NGFN-2 from the random sampling of cDNA libraries to the directed modelling, cloning and sequence validation of gene structures. This switching of strategy was done for mainly two reasons:
I) The number of human protein encoding genes is currently estimated to be around 25,000, more than half of them have known gene structures and full-length cDNAs are available. These cDNAs have mostly been cloned in three large-scale projects that are carried out worldwide, namely the Japanese Kazusa/NEDO project, the German cDNA Consortium, and the US Mammalian Gene Collection. While these projects have generated ESTs from over four million cDNAs, the novelty rate in random sampling has dropped dramatically over the years. This because abundant transcripts have been mostly cloned and the remaining genes and transcripts are represented in the cDNA libraries in too low numbers to allow for their detection by random sampling. II) The genomes of human, mouse, and several other model organisms have been sequenced. In conjunction with EST and full-length cDNA projects these sequence resources facilitate a comparative genome analysis that has greatly enhanced the capabilities of modelling gene structures in silico, and verifying these models through experimentation.
The directed cloning of ORFs from gene structures has the additional advantage that the resources can be immediately exploited through expression of the encoded proteins. Along this line, the sequence validation of Gateway Entry-clone resources of the cDNA Consortium forms the basis for almost any cellular functional gene analysis that is performed in SMP-Cell and beyond.
Large-scale cDNA resources
Within the German Genome Project and NGFN-1 the partners of the consortium have sequenced over 280,000 ESTs (145 Mb) and over 15,000 (50 Mb) full-length cDNAs (1). All clones were generated within the consortium (4). The sequences have been deposited in the EMBL/GenBank/DDBJ databases (Fig. 2), while the clones are distributed by the RZPD and are actively utilized in the community (see below).
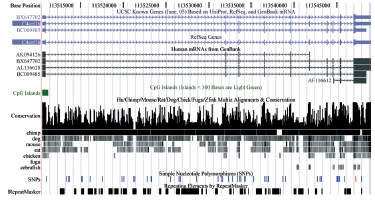 |
|
The SMP-Cell in the NGFN-2 builds on the cDNA resources generated in the DHGP and NGFN-1 projects, and has already established a resource of over 1,000 sequence validated Gateway Entry clones. Sequencing of entry clones was identified as an essential step in the cloning process to provide the experimental projects, where the ORFs are exploited at the cellular level, only with quality controlled and standardized expression constructs. Only the sequencing process could help to rule out errors that could have been introduced in the ORFs amplification and cloning processes, where frame shift mutations are the most deleterious. This type of mutation renders the respective clones not useful for downstream experimentation.
However base substitutions could as well have dramatic effects on the functionality of the encoded proteins and at least require detection and proper annotation. Validated clones in the end form a standardized and quality controlled resource for subsequent functional characterization of the encoded proteins.
Results from sequencing feed back into the ORF cloning project, as for instance error rates of DNA polymerases can be evaluated quantitatively and the optimal enzymes selected for amplification. Close ties with the infostructure project of the consortium and with the bioinformatics project of SMP-Cell are implemented to form a fast link between cloning, sequence validation, annotation, and exploitation of ORF-clones.
Exploitation of sequence verified resources
Researchers worldwide make use of the resources provided by the German cDNA Consortium (see http://www.genome.org/cgi/content/full/11/3/422 for citations of Reference (1)). Especially the partners within SMP-Cell utilize the resources in large scale protein localization (5), high-throughput cellular functional gene analysis (6, 7), and increasingly in single gene analysis and annotation (8-10). Thus far the initial cDNAs required amplification and subcloning to produce Gateway entry clones, now the SMP-Cell generates such entry clones directly. The increase in the throughput of cloning and sequence validation has already enhanced the potential of the experimental projects to screen a larger number of proteins for their disease relevance, and to identify candidate genes and proteins for further analysis.
International standing
The German cDNA Consortium has established a solid standing in the international scientific community, much of the reputation of the DHGP and now the NGFN is based on the visibility of the clones and sequences that have been generated by this consortium. The Consortium was one major provider of sequences to the international H-invitational initiative to annotate the human transcriptome (11). Over 100 researchers from 60 countries have assembled twice in Tokyo, aiming at a complete description of the human genes, splice forms and their sequence variation.
Outlook
With the switch from random sampling of cDNA libraries to the directed modelling, cloning, and sequence validation of ORF resources, the German cDNA Consortium has again taken a leading role in the international effort to provide the community with sequence validated full-length cDNA resources. Use of the Gateway system and the principle of selecting specifically the protein coding regions for cloning has greatly fostered the immediate applicability of these cDNAs. Sequence validation of all representative clones for every gene and splice variant, cloned in open and closed forms, generates a highly valuable resource for functional genomics. During NGFN-2 the sequencing capacities of the German cDNA Consortium will guarantee a tight QC of resources and lay the ground for a successful exploitation of the clone resources in SMP-Cell and beyond.
Lit.: 1. Wiemann S et al. Toward a Catalog of Human Genes and Proteins: Sequencing and Analysis of 500 Novel Complete Protein Coding Human cDNAs. Genome Res. 2001 Mar;11(3):422-35. 2. Wiemann S et al. The German cDNA Network: cDNAs, functional genomics and proteomics. Journal of Structural and Functional Genomics. 2003 4(2-3):87-96. 3. Wiemann S et al. cDNAs in functional genomics and proteomics: The German cDNA Consortium. CRBiologies. 2003 326:1003-9. 4. Wellenreuther R et al. SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones. BMC Genomics. 2004 5:36. 5. Simpson JC et al. Systematic subcellular localization of novel proteins identified by large scale cDNA sequencing. EMBO Rep. 2000 1(3):287-92. 6. Starkuviene V et al. High-content screening microscopy identifies novel proteins with a putative role in secretory membrane traffic. Genome Res. 2004 Oct;14(10):1948-56. 7. Arlt DH et al. Functional profiling: from microarrays via cell-based assays to novel tumor relevant modulators of the cell cycle. Cancer Res. 2005 65(17):in press. 8. Neubrand VE et al. Gamma-BAR, a novel AP-1 interacting protein involved in post-Golgi trafficking. EMBO J. 2005 24:1122-33. 9. Fleischer S et al. PML-Associated Repressor of Transcription (PAROT), a Novel KRAB-ZINC Finger Repressor is Regulated through Association with PML Nuclear Bodies. J Biol Chem. 2005:submitted. 10. Wiemann S et al. Alternative pre-mRNA processing regulates cell-type specific expression of the IL4l1 and NUP62 genes. BMC Biol. 2005 Jul 19;3(1):16. 11. Imanishi T et al. Integrative Annotation of 21,037 Human Genes Validated by Full-Length cDNA Clones. PLoS Biol. 2004 Apr;2(6):856-75.
Please visit the Project's Website


