Introduction
Malignant gliomas are believed to derive from astrocytic precursor cells (anaplastic astrocytomas and glioblastomas) or from ependymal cells lining the ventricular system (anaplastic ependymomas). High-grade astrocytomas represent 6.6 % of pediatric brain tumors. Similarly, ependymomas constitute 10 % of brain tumors in children. However, they represent more than 50% of brain tumors arising in children younger than 5 years. Although the survival rates have increased during the last years especially due to better surgical techniques and postoperative treatment the prognosis is still worse compared with other childhood cancers. Survivors frequently suffer from severe central nervous late effects after multimodal intense treatment. In addition, irradiation and chemotherapy significantly increase the risk of second cancer. The biological behaviour of ependymomas is difficult to predict based on clinical and histopathological factors. Therefore, the identification of genetic markers with predictive value would be very helpful.
Genetic analysis has shown that both ependymal and atrocytic anaplastic tumors of childhood seem to follow genetic pathways different from those of the corresponding adult cancers (Rickert et al, 2001; Dyer et al., 2002). Therefore, it would be very interesting to directly analyze and compare anaplastic gliomas from adults and children.
In both, anaplastic astrocytomas and ependymomas, gains of chromosomal material of chromosome 1q have been identified to be associated with worse prognosis. The identification of the genes responsible for this behaviour may result in new insights in the biology of malignant gliomas of childhood and may lead to the definition of novel therapeutic targets.
Results / Project Status
Genetic analysis of candidate genes
In initial studies, we screened ependymomas for allelic losses of chromosomal arms 6q, 13q, 17p, and 22q. On 22q, we were able to define a region of loss distinct from NF2 (Kraus et al., 2001), which is frequently affected in adult patients with spinal ependymomas (Ebert et al., 1999). We were also able to exclude a major role of SV40, JC or BK infection in the development of ependymomas (Weggen et al., 2000). On chromosome 17p we identified the HIC1 tumor suppressor gene to be frequently silenced in medulloblastoma and ependymomas (Waha et al., 2003, 2004). Most of the high-grade astrocytoma samples have already been characterized for the status of TP53, PTEN, EGFR, and MDM2 (Kraus et al., 2002).
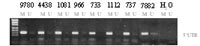
Silencing of HIC-1 tumor suppressor gene by methylation in human ependymomas
Ependymomas are among the most common brain tumors in children. They develop from ependymal cells lining the ventricular system of the CNS. Previous studies have demonstrated a significant rate of allelic loss at chromosome 17p13.3. The HIC-1 putative tumor-suppressor gene, which exhibits hypermethylation and loss of expression in various tumor entities including medulloblastomas and gliomas, maps to the affected region. In the present study, we analyzed HIC-1 in ependymomas. Therefore, we applied methylation-specific PCR of the 5'-untranslated region as well as of a central region of HIC-1 and bisulfite sequencing to determine the methylation status in 52 ependymomas of different histologic subtypes, grades and locations. In addition, we used a competitive RT-PCR approach for sensitive assessment of HIC-1 transcripts. Hypermethylation
of at least one of the 2 analyzed regions was found in 43/52 (83%) cases. There was a significant correlation between hypermethylation of HIC-1 and nonspinal localization (p = 0.019) as well as age. Of 27 ependymomas, 22 (81%) showed absent or low expression of HIC-1. The elevated methylation of HIC-1 in nonspinal ependymomas supports the hypothesis that spinal and nonspinal ependymomas represent genetically distinct entities.
Genome-wide screening of copy number changes and epigenetic alterations
High grade gliomas of childhood are analysed by matrix CGH. Tumour DNA are currently hybridized against arrays of clones that cover the whole genome. These arrays have been developed by the group of Prof. Dr. P. Lichter at the Department of Molecular Genetics at the DKFZ. Analysis will be performed with DNA from 20 high grade astrocytomas and 30 ependymomas. The findings will be compared with the data generated from adult high grade gliomas. We will validate recurrent copy number changes by microsatellite analysis and duplex-PCR with fluorescent primers. Products are quantified on a ABI 377 genetic analyser. Identical tumor DNA samples will be analysed for epigenetic changes by differential methylation analysis (DMH). Recurrent hypermethylation of CpG islands will be verified by methylation-specific PCR.
Prognostic relevance
The presence or absance of recurrent genetic alterations will be tested in regard to an association to a good vs. worse outcome of the patients. Known prognostic factors such as location, age and extent of surgical removal are taken into account.
Identification of a commonly altered region on chromosome arm 1q and identification of candidate genes
We will try to define minimal regions affected by dosage gains in high grade gliomas of childhood. We will try to find out if in astrocytic and ependymal tumors an identical region is affected. If cases with smaller regions of gains are identified then the borders of the amplified regions will be determined by region specific BAC arrays and PCR analyses. In the case that this strategy will be sufficient to narrow down a region the expression levels of genes located here will be analysed. Genes affected by amplification or overexpression will be genetically and functionally analysed. For mutational analysis, SSCP screening and sequence analysis will be performed. For functional studies, cDNA will be cloned into appropriate expression vectors for overexpression studies, and RNAi technology used for knock-down experiments. If antibodies are available against their products they will be used for immunohistochemistry of sections.

Outlook
The systematic analysis of genetic and epigenetic changes in high grade gliomas of childhood will lead to the identification of alterations with predictive value. In future clinical trials, such markers can be prospectively analysed and may serve as tools for stratification of patients according to their individual risk. In addition, the detailed study of gain of genetic material of chromosomal arm 1q will lead to the identification of functionally important genes located there. Comparison between adult and childhood high grade gliomas may identify specific genetic signatures for childhood high grade brain tumors which may help to understand their biology.
Lit.: 1. Dyer, S., Prebble, E., Davison, V., Davies, P., Ramani, P., Ellison, D., Grundy, R. Genomic imbalances in pediatric intracranial ependymomas define clinically relevant groups. Am. J. Pathol. 2002: 161: 2133-41. 2. Rickert, C.H., Sträter, R., Kaatsch, P., Wassmann, H., Jürgens, H., Dockhorn-Dworniczak, B, Paulus, W. Pediatric high-grade astrocytomas show chromosomal imbalances distinct from adult cases. Am. J. Pathol. 2001: 158: 1525-32. 3. Ebert, C., von Haken, M., Meyer-Puttlitz, B., Wiestler, O.D., Reifenberger, G., Pietsch, T., von Deimling-A. NF2 mutations and allelic loss of chromosome 22q in ependymal tumors: Association with intramedullary spinal localization. Am. J. Pathol. 1999, 155: 627-32. 4. Timmermann, B., Kortmann, R.D., Kühl, J., Meisner, C., Slavc, I., Pietsch, T., Bamberg. M. Combined postoperative irradiation and chemotherapy for anaplastic ependymomas in childhood: results of the German prospective trials HIT 88/89 and HIT 91. Int. J. Radiat. Oncol. Biol. Phys. 2000, 46: 287-95. 5. Weggen, S., Bayer, T.A., von Deimling, A., Reifenberger, G., von Schweinitz, D., Wiestler, O.D., Pietsch, T. Low frequency of SV40, JC and BK polyomavirus sequences in human medulloblastomas, meningiomas and ependymomas. Brain Pathol. 2000, 10: 85-92. 6. Kraus, J.A., de Millas, W., Sörensen, N., Herbold, C., Schichor, C., Tonn, J.C., Wiestler, O.D., von Deimling, A., Pietsch, T. Evidence for a tumor suppressor gene at 22q11-q12 involved in the pathogenesis of human ependymomas and distinct from hSNF5/INI1. Acta Neuropathol. 2001, 102: 69-74. 7. Kraus, J.A., Felsberg, J., Tonn, J.C., Reifenberger, G., Pietsch, T. Molecular genetic analysis of the TP53, PTEN, CDKN2A, EGFR, CDK4 and MDM2 tumor-associated genes in supratentorial primitive neuroectodermal tumors and glioblastomas of childhood. Neuropathol. Applied Neurobiol. 2002, 28: 325-33. 8. Waha, A., Waha, A., Koch, A., Meyer-Puttlitz, B., Weggen, S., Sörensen, N., Tonn, J.C., Albrecht, S., Goodyer, C.G., Berthold, F., Wiestler, O.D., Pietsch, T. Epigenetic silencing of the HIC-1 gene in human medulloblastomas. J Neuropathol Exp Neurol 2003, 62:1192-201.9. Waha, A., Koch, A., Mack, H., Tonn, J. C., Sörensen, N., Berthold, F., Wiestler, O. D., Pietsch, T. and Waha, A. Analysis of HIC-1 methylation and transcription in human ependymomas. Int J Cancer 2004;110: 542-9. 10. Pietsch, T., Taylor, M.D., Rutka, J.T. Molecular pathogenesis of childhood brain tumors. J Neurooncol. 2004;70:203-15. 11. Waha, A., Guntner, S., Huang, T.H., Yan, P.S., Arslan, B., Pietsch, T., Wiestler, O.D., Waha, A.Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005; 7:193-9.


