Introduction
Genetic instability is a hallmark of neoplasia and is thought to be instrumental in cancer initiation and progression. Numerous techniques have been developed for the mapping of chromosomal aberrations, but the identification of candidate genes has been hindered by their low resolution and limited throughput capacity. A possible solution to this predicament emerged with the development of matrix-CGH, a chip-based method for the genome-wide profiling of DNA-copy number status. In matrix-CGH, DNA from a test (e.g., tumor) and a reference genome (genomic DNA from a normal individual) are differentially labeled and hybridized to an array of large-insert genomic clones (usually BAC, PAC or cosmid) of known map position. The fluorescence ratios of individual clones provide information about the relative copy number of the corresponding sequences in the test and reference genomes. The resolution and sensitivity of matrix-CGH are determined by the size and mapping position of the clones on the array.
Application of genome-wide matrix-CGH in different tumors is revealing that selection for specific genes relevant to tumor pathogenesis as well as effects of specific types of genetic instability shape tumor genomic profiles. This causal, non-random relationship of genomic profile and tumor pathomechanism offers a unique opportunity to address important clinical questions such as differentiation of tumor subgroups with different clinical behavior in regard to prognosis, probability of progression, and therapy response. Furthermore, recurrent deletions or amplifications of genomic material usually are good indicators of the presence of tumor suppressor or oncogenes in the imbalanced chromosomal region. Fine-mapping of the affected regions using high-resolution matrix-CGH arrays will facilitate the identification of new target genes for therapeutic interference, especially when combined with the analysis of DNA methylation status, gene- and protein expression.
Well-known examples of diagnosis and treatment decisions based on analysis of genetic aberrations include Herceptin therapy of breast cancer patients with ERBB2 amplification, and the use of STI 571 (Glivec) for tumors with Bcr-Abl translocation or c-Kit mutation. It seems likely that many more such markers will find entry into clinical applications in the future. Genomic DNA has high chemical stability, and its copy number does not vary over time or under different physiological conditions. This makes centralized and standardized DNA-based processing and diagnosis of tumor biopsies a realistic option, in particular since molecular methods to handle genomic DNA already are well established in many clinical laboratories.
Results
CDK6 is a prognostic marker in medulloblastoma
We performed genome-wide analysis of DNA-copy number imbalances in 47 medulloblastomas using comparative genomic hybridization to large insert DNA microarrays (matrix-CGH). RNA and protein expression of selected candidate genes identified by matrix-CGH were analyzed by RQ-PCR and immunohistochemically on tissue microarrays representing medulloblastomas from 189 clinically well-documented patients, respectively. To identify novel prognostic markers, genomic findings and protein expression data were correlated to patient survival. Matrix-CGH analysis revealed frequent DNA-copy number alterations of several novel candidate regions. Among these, gains at 17q23.2-qter (p<0.01) and losses at 17p13.1-13.3 (p=0.04) were significantly correlated to poor prognosis. Within two of the most frequently gained chromosomal regions, 17q23.2-ter and 7q21.2, confined amplicons were identified that contained the PPM1D and CDK6 (Fig.1) genes, respectively.

Immunohistochemistry revealed strong expression of PPM1D in 148 of 168 (88%) and CDK6 in 50 of 169 (30%) medulloblastomas. Overexpression of CDK6 significantly correlated with poor prognosis (p<0.01) and represented an independent prognostic marker of overall survival on uni- (Fig. 2) and multivariate analysis (p=0.02).
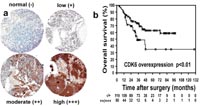
CDK6 overexpression represents a novel molecular marker that can be analyzed by immunohistochemistry on routinely processed tissue specimens and may facilitate the prognostic assessment of medulloblastoma patients. Furthermore, PPM1D and CDK6 may link the TP53 and RB1 tumor suppressor pathways to medulloblastoma pathomechanisms.
i(17q) breakpoints in medulloblastoma
The most frequent chromosomal rearrangement in medulloblastoma is isochromosome 17, or i(17q). It is detected in up to 40% of tumors, suggesting that it serves an important role in medulloblastoma pathogenesis, possibly mediated by the disruption or permanent activation of a gene at the breakpoint. To address this question, we performed a detailed analysis of chromosome 17 DNA-copy number from 18 medulloblastomas previously shown to carry an apparent i(17q). We identified two breakpoint regions, one well within band 17p11.2 (n=16) and a second within the pericentromeric region (n=2). In order to map the breakpoints more precisely, we constructed a tiling-path matrix-CGH array covering chromosomal band 17p11.2 to the centromere and utilized it to delineate two small breakpoint intervals in seven of the medulloblastomas and nine hematological neoplasias with i(17q) (Fig. 3). The former contains two breakpoint clusters that each co localize with a pair of head-to-head inverted DNA sequence repeats, and the latter maps close to a region of #-satellite repeats. No consensus coding sequence localize in these regions. Together, these data strongly suggest that effects of i(17q) in medulloblastoma pathogenesis are mediated by gene dosage effects of genes on 17p or 17q rather than the disruption or deregulation of a “breakpoint gene”. Furthermore, we identify artifacts introduced in DNA-copy number data by cross-hybridization of low copy repeat sequences, thereby demonstrating the problems these can pose to the interpretation of diagnostic microarrays.

DNA copy number profiling of gliomas
We have completed DNA-copy number profiling of the 96 primary gliomas constituting the “Glioma Core Collection” of the NGFN2 Brain Tumor Network. For this purpose, a 6000 clone matrix-CGH chip developed during NGFN1 for genome-wide tumorgenetic screening was augmented for the analyses of brain tumors by adding 2000 additional clones. We currently are concluding primary data analyses. Furthermore, we are in the process of screening 50 samples of aggressive childhood brain tumors.
Outlook
Our work on medulloblastoma demonstrates that matrix-CGH is a powerful technique capable of identifying clinically relevant molecular markers and putative therapy target genes. The potential of the method can further be enhanced by parallel and comparative profiling of DNA methylation, RNA and protein expression. In co-operation with our partners in the NGFN2 Brain Tumor Network we will continue to pursue this type of integrative approach to brain tumor research.
Lit.: 1. Mendrzyk F et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. JClinOncology 2005, in revision. 2. Mendrzyk F et al. The isochromosome breakpoints on 17p in medulloblastoma are flanked by different classes of DNA sequence repeats. submitted.


