Introduction
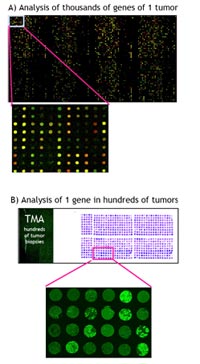
A major aim within the glioma network is the identification of tumor-specific genetic aberrations. On the one hand, this provides important new insights into the molecular mechanisms underlying tumor development, on the other hand the results could have important impacts regarding clinical diagnostic and therapy. Genome-wide screening approaches such as matrix-CGH and expression profiling are used in many of the subprojects described to detect genetic aberrations. However, frequently only limited numbers of cases of a given tumor entity (usually ~ 50) are analyzed by DNA-chip technologies for time and cost reasons. Therefore the pathogenetic and clinical relevance of candidate markers needs to be further assessed on larger tumor cohorts. For this tissue microarrays (TMAs) can be efficiently used.
TMAs represent ordered arrays of tissue cores on glass slides, which can be analyzed by fluorescence in situ hybridization (FISH) or immunohistochemisty (IHC). They provide rapid information, whether genomic changes or aberrant protein expression are associated with clinical parameters, such as grading, staging or overall survival (OS). The combined approach of genome-wide screening and TMA analysis is illustrated in Fig. 1. Beside malignancies of the central nervous system (see below), we performed TMA analyses of various other tumor entities including squamous cell carcinoma of the head and neck (1-3), adenoid cystic carcinoma of the salivary gland (4,5), colorectal carcinoma (6), as well as different types of malignant lymphoma (7).
Results/Project Status
TMA analysis of Medulloblastoma:
Medulloblastoma represents one of the most common central nervous system malignancies of childhood. For our studies we generated a TMA composed of 189 medulloblastoma tissue cores. All diagnoses were confirmed by histological assessment of specimens obtained at surgery by at least two neuropathologists according to the criteria of the World Health Organization (WHO) classification.
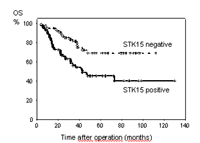
A major interest was to identify genes that predict for overall survival (OS). An initial gene expression profiling analysis of 35 cases revealed 54 genes which were possibly associated with unfavourable clinical course. To analyze the nine most promising candidate genes in more detail, a TMA composed of 180 medulloblastoma tissue cores was generated for IHC and FISH experiments. In line with the gene expression profiling analysis, a positive staining for STK15, stathmin 1, and cyclin D1 was found associated with unfavourable OS. Compared with clinical parameters such as gender, age, metastatic stage, extent of tumor resection, application of chemotherapy, and tumor grade, positive staining for STK15 was identified as an independent prognostic factor for OS (Fig. 2). Moreover, additional gene copy numbers of MYC and STK15 predicted for poor survival. This study demonstrated, that the combined approach of gene expression profiling and tissue microarray analysis is highly suitable for identification of genes predicting for survival. In addition, we showed, that genes associated with cell proliferation (cyclin D1), transcription (MYC), and especially mitosis (stathmin 1, STK15) appear to play a crucial role in medulloblastoma pathogenesis.
Within the NGFN2 glioma network we currently perform further analyses of the same medulloblastoma TMA in order to define the clinical role of genes, which were selected based on matrix-CGH-CGH results (collaboration with B. Radlwimmer and P. Lichter, Heidelberg). All of them are located within distinct chromosomal subregions recurrently amplified in medulloblastoma. Up to now, overexpression of a gene product, which is involved in cell cycle control, has been demonstrated to be an independent prognostic marker of OS (Mendrzyk et al. submitted). Another study deals with the characterization of chromosomal breakpoints on isochromosome 17, which represents the most frequent chromosomal rearrangement in medulloblastoma. For this, bacterial artificial chromosome (BAC) clones flanking the breakpoint region are used for FISH experiments on TMAs (Mendrzyk et al. submitted).
TMA analyis of Intracranial Ependymoma:
Intracranial ependymoma is another common CNS tumor of childhood. We generated a TMA of 170 patients with newly diagnosed intracranial ependymomas. Pathological diagnosis was made according to the WHO classification. From all patients follow-up data were available. Based on relatively small tumor collections, various studies have suggested, that numerical chromosomal changes are associated with an unfavourable clinical course. Furthermore, recent matrix-CGH analyses from the glioma network (B. Radlwimmer, F. Mendrzyk and P. Lichter) revealed distinct and frequently rearranged chromosomal subregions spanning putative oncogenes. The most interesting genomic regions are currently analyzed by FISH in order to verify the prognostic significance of these aberrations in a representative tumor collection.
TMA analysis of Oligodendroglioma and Astrocytoma (Grade II and III):
The histologic classification of gliomas is based on oligodendroglial- or astrocytic-like morphology. Gliomas are distinguished in astrocytic tumors (grade I, pilocytic astrocytoma to grade 4, glioblastoma), oligodendroglial tumors and mixed tumors (grade II and III). Mixed glioma (oligoastrocytoma) contains both, neoplastic astrocytes as well as neoplastic oligodendroglial cells. Genetic aberrations play an increasing role in routine clinical diagnosis of these tumors. For example, it has been found, that combined LOH on chromosomal arms 1p and 19q is a hallmark for oligodendroglioma, and has emerged as prognostic marker of longer survival and better chemotherapeutic response. The target genes within these chromosomal regions are still unknown.
In collaboration with Ch. Herold-Mende (Heidelberg) and G. Reifenberger (Düsseldorf) from the glioma network, a TMA with 240 different core biopsies (oligodendrogliomas, astrocytomas and oligoastrocytomas) from 123 different patients was constructed. From all tumors detailed clinical data are available including therapy modalities and clinical course. Different studies are currently performed using this TMA. One of them is related to the identification of the chromosome 1p/19q tumor suppressorgenes (see above). By applying FISH using 1p and 19q-specific clones, we try to narrow down the minimally overlapping region, which is consistently deleted in oligodendroglioma.
Another project is concerned with the expression of MGMT (i.e. O6-methy-guanin-DNA-methyltransferase). The MGMT gene has been frequently found inactivated (e.g. by promoter methylation) in CNS tumors resulting in the lack of an important DNA-repair function. This might become very important in the context of therapy decisions, since MGMT-negative tumors are expected to have a higher probability to respond to alkylating cytostatic drugs, whereas MGMT-positive tumors should be rather resistant to chemotherapy. We currently investigate the MGMT-expression status in our tumor collection and correlate the results with therapy response data. In this way we hope to get further support for MGMT as a suitable marker for therapy stratification in these tumors.
Finally, in a collaboration with W. Roth, P. Krammer and H. Walczak (Heidelberg), we investigate, whether antibodies against apoptotic or anti-apoptotic proteins are associated with clinical parameters. Antibodies, which where tested up to now, included TRAIL-R1, TRAIL-R2, TRAIL-R3, TRAIL-R4 as well as c-Flip.
Outlook
Within the glioma network we are on the way to generate two more TMAs, one Glioblastoma/Astrocytoma-TMA with about 700 tumors (in collaboration with Ch. Herold-Mende, Heidelberg, and G. Reifenberger, Düsseldorf). The second TMA (NOA-04 TMA) is produced in close collaboration with W. Wick and M. Weller (Tübingen) as well as Th. Pietsch (Bonn) and contains tumor biopsies from about 300 patients with anaplastic glioma. These tumors are part of a randomized phase III clinical study initiated by the German Neurooncology Working Group (NOA) of the German Cancer Society. The study was designed to investigate, whether predictions can be made with regard to chemotherapy and radiotherapy response.
Lit.: 1. Freier K et al. Tissue microarray analysis reveals site-specific prevalence of onogene amplifications in head and neck squamous cell carcinoma. Cancer Res. 2003, Mar 15;63(6): 1179-82. 2. Freier K et al. Distinct site-specific oncoprotein overexpression in head and neck squamous cell carcinoma: A tissue microarray analysis. Anticancer Res. 2003; Sep-Oct 23(5A):3971-7. 3. Sticht C et al. Amplification of CyclinL1 is associated with lymph node metastases in head and neck squamous cell carcinoma (HNSCC). Br J Cancer. 2005 Feb 28;92(4):770-4. 4. Freier K et al. Novel copy number gains on chromosome 22q13 in adenoid cystic carcinoma (ACC) of the salivary gland revealed by comparative genomic hybridization (CGH) and tissue microarray (TMA) analysis Cytogenetics and Cell Genetics, 2005 May;159(1):89-95. 5. Freier K et al. Differential KIT protein expression in histological subtypes of adenoid cystic carcinoma (ACC) of the salivary gland. Oral Oncology 2005, Jul 26; Epub ahead of print. 6. Völp K et al. Expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the anti-apoptotic c-IAP2 protein in human colon carcinomas. Gut, 2005, in press.7. Mathas S et al. Elevated Bcl-3 expression and p50 complex formation in anaplastic large cell, classical Hodgkin- and peripheral T cell lymphomas, Blood, 2005, in press. 8. Neben K et al. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res. 2004 May 1;64(9):3103-11.


