Introduction
Brain tumors are diverse and range widely in their clinical presentation, biological aggressiveness, histological differentiation, and response to therapy. The most common type of primary malignant brain tumors are gliomas. They can occur at any age but most frequently affect elderly people. The diffusely invasive properties of malignant gliomas make them nearly impossible to totally resect. After surgical resection radiation therapy as well as chemotherapy are state of the art treatments. Nevertheless recurrent growth occurs in most cases of malignant glioma. In particular, highly aggressive, rapidly growing tumors are exposed to hypoxia or even anoxia, which occurs as a consequence of inadequate blood supply. But also therapeutic measures like irradiation give rise to hypoxic conditions in tumor cells. The presence of hypoxia in tumors has a negative impact on patient prognosis. Both hypoxia and consecutive hypoxia/reoxygenation exert a variety of influences on tumor cell biology. Among these are activation of certain signal transduction pathways and gene regulatory mechanisms, adapting hypoxic tumor cells to an anaerobic environment by the transcriptional induction of genes involved in glycolysis, hematopoieses, angiogenesis, apoptosis and tissue invasion. Most of these mechanisms contribute to tumor progression. Moreover hypoxic tumor cells are resistant to conventional chemotherapy and radiotherapy and thus hypoxia promotes a more malignant phenotype. The protein hypoxia-inducible factor-1 (HIF-1) mediates many of these changes in gene expression. In response to oxygen availability posttranslational modifications such as prolyl-hydroxylation, ubiquitination, sumoylation, and acetylation are significant modifiers for HIF-1alpha activity. A recent report has shown that SUMO-1 expression is up-regulated in response to hypoxia in adult mouse brain and heart in vivo (1). It could be shown, that SUMO-1 co-localizes and co-immunoprecipitates with HIF-1alpha in the nucleus. SUMO (small ubiquitin-related modifier) represents a class of ubiquitin-like proteins which are conjugated posttranslationally, like ubiquitin, by a set of ligases to cellular regulatory proteins, including oncogenes and tumor suppressor genes, that play key roles in the control of cell growth, differentiation and apoptosis. SUMO conjugation affects subcellular localization and stability of its substrates as well as transcriptional activities. Given the substrates involved, protein sumoylation is expected to be important in the course of tumor genesis and, accordingly, altered in human cancer.
Project Status
Comparative genomic hybridization and gene expression analysis cover the genomic and transcriptional alterations in a tumor; however, they are not able to reflect translational alterations of these tumors. Extending the studies carried out on the DNA and RNA level to the protein level will allow the comparison and correlation of data from gene expression analysis and RNA expression profiling with changes in protein expression. In close cooperation with the group of Weller/Wick and as a complementary approach to gene expression profiling performed by Weller/Wick and Hahn/Lichter we therefore aim to investigate changes in the overall protein pattern as well as in the sumoylation pattern of malignant glioma cell lines after hypoxia treatment compared to normoxic controls utilizing most-advanced proteomics techniques.
We first focused our studies on differences in the sumoylation pattern of glioma cells cultivated under hypoxic conditions. To achieve this goal, we produced specific antibodies against the various proteins of the SUMO family. To specifically detect SUMO-1, -2 and -3 proteins, polyclonal antibodies were raised by immunization of guinea pigs with 5 different peptides (2 animals/peptide) in collaboration with PSL GmbH, Heidelberg, Germany. The peptides were selected on the basis of differences in the primary sequence of SUMO-1, -2 and -3, predicted antigenicity and surface probability.
The following peptides were synthesized:
SUMO1-N-terminal (NT) # MSDQEAKPSTEDLC
SUMO1-internal (INT) # CEGQRIADNHTPKE
SUMO2-N-terminal (NT) # MADEKPKEC
SUMO2/3-internal (INT) # CRFRDGQPINETDTPA
SUMO3-C-terminal (CT) # CGVPESSLAGHSF
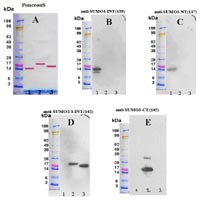
Cysteine (C) residues were added to the N- or C-terminus, respectively, to achieve covalent binding to the carrier protein KLH (keyhole limpet hemocyanine) prior to immunization. The antisera obtained from the immunizations were purified on peptide affinity columns and the specificities of the polyclonal antibodies were tested on immunoblots using ovalbumin-coupled peptides as well as the corresponding recombinant proteins. By this approach we obtained specific antisera against SUMO-1 and SUMO-3 and one antiserum that recognizes SUMO-2 and -3 (Figure 1). Due to the high sequence homology between SUMO-2 and SUMO-3, we did not obtain a specific antibody against SUMO-2.
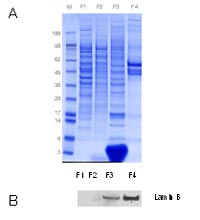
A feature of SUMO modification is, that for most substrates, only a small portion of is sumoylated at a given time. To beable to detect low abundant sumoylated proteins we have to reduce the complexity of protein samples by analyzing subcellular structures, in particular the nuclear fraction. To obtain subcellular fragmentation of flash frozen cell pellets and later of flash frozen fragmented tissues, we decided to utilize the subcellular proteome extraction kit which is commercially available from Calbiochem (Merck Biosciences, Darmstadt, Germany). This kit was tested with flash-frozen cells from a benign prostate hyperplasia (BPH-1) cell line as shown in Figure 2.
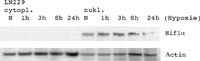
To inhibit the action of cysteine proteases which are known to degrade sumoylated proteins very rapidly, the extraction buffers were supplemented with 10mM iodoacetamide and 10mM N-ethylmaleimide. As starting material for the identification of hypoxia-associated proteins and modifications we used the LN229 Glioma cell line provided by our co-operation partners.
Cells were maintained and exposed to hypoxia at five different time-points (zero, 1hour, 3hours, 8hours and 24hours) Aliquots of nuclear and cytoplasmic extracts obtained at different time-points were precipitated and analyzed for HIF-1alpha.
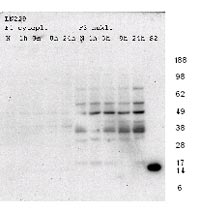
In a first experiment to detect sumoylated proteins in our extracts, cytoplasmic and nuclear fractions were separated by 1D-gel electrophoresis and blotted onto a PVDF membrane. After probing with an antibody specific for SUMO-2 and SUMO-3 protein immunopositive signals could be detected in a molecular range between 20kDa and 80kDa (Figure 4).
In future experiments we will investigate in more detail changes in the overall protein pattern and in the pattern of sumoylated proteins from nuclear cell extracts at different time points of hypoxia treatment. Cell extracts will be separated by high-resolution 2 dimensional (2D) gel electrophoresis (Figure 5).
For the analysis of changes in the protein pattern protein spots will be visualized either by labeling with fluorescent dyes prior to the electrophoretic separation or by using a silver stain protocol for analytical gels compatible with mass spectrometry. Computer-assisted image analysis of the gels will allow the detection of differentially expressed proteins (Figure 6).


Proteins spots that are recognized by image analysis to be differentially up- or down-regulated at least 4fold will be identified by MALDI peptide mass fingerprinting and/or nanoLC-ESI-MS/MS sequencing. Database search will be performed against protein-, EST- and genomic databases. Protein expression data from our studies will be compared with gene expression data obtained from the same set of cell lines (in collaboration with Weller/Wick and Hahn/Lichter).
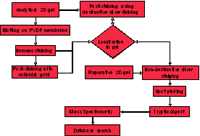
For the detection of differences in the sumoylation pattern of nuclear proteins and for the identification of the sumoylated proteins by mass spectrometry the above mentioned approach has to be extended. The experimental procedure is outlined in Figure 7. Analytical 2D gels representing different time points of hypoxia treatment will be blotted to PVDF membrane and immunostained with the respective SUMO antibodies to detect sumoylated proteins. The corresponding proteins spots on analytical or preparative 2D gels which were run in parallel will be localized, excised from the gel and finally identified as described above.
The Protein Analysis Facility of DKFZ provides outstanding experience in proteomics analysis of biological systems. One major focus of the group is the identification of differentially expressed proteins associated with the development of chemo- and thermoresistance in melanoma (2), gastric, and pancreatic carcinoma cell lines (3). Furthermore, the group has long-standing experience in the identification of proteins and protein-protein interactions and the characterization of posttranslational modifications. Within the funding period of NGFN-1 almost 3000 cancer-related proteins from partners in the core area platform as well as from external partners and partners in the disease-oriented networks could be successfully identified and characterized. The group is excellently equipped and has established a variety of mass spectrometric techniques including MALDI-MS and MALDI-PSD as well as ESI-MS/MS with and without LC separation. The current instrumentation consists of two MALDI-TOF mass spectrometers, one ESI ion-trap mass spectrometer and one ESI-Q-Tof mass spectrometer both coupled with a nanoHPLC system including autosampler. For database searches, a 4-processor Linux cluster was recently established. In addition, the group has successfully set up a high-resolution 2D gel electrophoresis system including image analysis and data mining software for complex proteomics studies.
Outlook
Immunhistochemistry will be used to evaluate potential protein markers associated with hypoxia. Commercially available antibodies as well as peptide-specific antisera, which will be generated by immunization of guinea pigs with KLH coupled peptides, will be tested on tissue microarryas (e.g. NOA-04 tissue microarray) in collaboration with the groups of Weller/Wick and Joos. In collaboration with the group of Eils the data will be transferred to the iChip-data base of the Bioinformatics Department of the DKFZ and subjected to refined analysis
Lit.: 1. Shao R et al. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo, FEBS Lett. 569 (2004) 293–300. 2. Sinha P et al. Identification of novel proteins associated with the development of chemoresistance in malignant melanoma using twodimensional electrophoresis. Electrophoresis. 2000 Aug;21(14):3048-57. 3. Poland J et al. Study of the development of thermoresistance in human pancreatic carcinoma cell lines using proteome analysis. Electrophoresis. 2004 Jan;25(1):173-83.


