Introduction
We aim at the development of bioinformatics methods for advancing the understanding of disease processes and the effect of genetic variations on protein function and drug therapies. To this end, application studies on structure and function prediction of medically relevant proteins are being conducted in cooperation with research groups from medical institutes. The bioinformatics service provided consists of various computational analyses of protein sequences of medical interest. Our findings have already led to plausible biological hypotheses that have been and are still of help in prioritizing further experiments. They have advanced the molecular understanding of impaired cellular mechanisms involved with specific diseases. In the following, we detail some of our bioinformatics studies.
Results
Sequence variations in the homologous products NALP3 and NOD2 of the genes CIAS1 and CARD15, respectively, have been associated with several autoinflammatory diseases (Fig. 1) that, although clinically different, share a similar inflammatory pathophysiology [1]. Both cytoplasmic proteins belong to a novel family of intracellular pathogen-sensing proteins that regulate the innate immune system and are known as NLRs (NACHT-LRR receptors) in analogy to the functionally and evolutionarily related transmembrane Toll-like receptors (TLRs). NALP3 is linked to chronic infantile neurological cutaneous and articular syndrome (CINCA, also known as NOMID), familial cold urticaria (FCU, also known as FCAS), and Muckle-Wells syndrome (MWS). NOD2 confers susceptibility to Blau syndrome (BS, also known as ACUG), early-onset sarcoidosis (EOS), and Crohn disease (CD), a chronic inflammatory bowel disease (IBD) [2]. NALP3 and NOD2 are both involved in the recognition of bacterial muropeptides and the regulation of inflammatory immune responses. The comparative analysis of genetic variations with respect to their structural impact on the protein level can afford important insights into disease mechanisms.
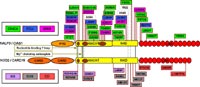
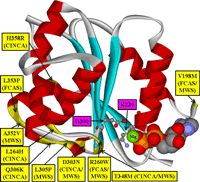
We assembled multiple sequence alignments of many NLRs sharing homologous domain architecture with N-terminal effector-binding domains (CARD or PYD domains), a central nucleotide-binding domain regulating signal transduction by conformational changes, and a C-terminal LRR (leucine-rich repeat) sensor domain [3]. Our sequence alignments and 3D structural models of the ATPase domain demonstrate that most of the disease-associated variants are located in highly conserved and spatially adjacent regions of the nucleotide-binding domain and possibly impair ATP hydrolysis and structural movements of protein domains (Fig. 2).
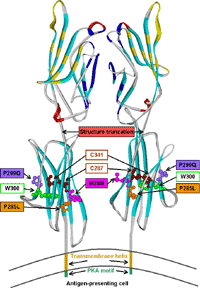
Furthermore, we could contribute to the structural analysis of the membrane-associated guanylate kinase DLG5, which has been associated with IBD like CARD15 [4]. We also predicted the structure of interferon-inducible IFI-200 proteins, which appear to contain two tandem DNA-binding OB folds at their C-termini [5]. This provided the long-sought explanation for their functional roles in transcriptional regulation implicated in inflammation and cancer. Most recently, our structural model of the BTNL2 gene product (Fig. 3), a butyrophilin-like member of the immunoglobulin superfamily, supported experimental investigations on the truncating splice site mutation associated with the multisystemic immune disorder sarcoidosis [6].
Outlook
An intricate cross-talk between complex signaling pathways that are induced by the recognition of pathogenic molecules is being discovered to mediate innate immunity. Surveillance receptors like NLRs, TLRs, and CLRs (C-type lectin-like receptors) trigger tailored gene expression profiles by the activation of important transcription factors such as NF-#B. Responsive genes range from proinflammatory cytokines and interferons to co-stimulatory molecules, which mount an immune response resulting in the removal and destruction of the invading pathogen.
Therefore, the impairment of essential signaling cascades by mutant proteins can lead to the dysregulation of human immunity, causing acute or chronic diseases of autoimmunity or immunodeficiency. However, the modification of innate immune responses by therapeutic targeting of the molecular processes may provide new opportunities for the clinical treatment of patients.
To support experimental research and planning, we are developing comprehensive online databases and web visualization interfaces on disease-associated proteins, identified sequence variants, and interaction networks. Our system will integrate the large amount of biomedical literature and experimental results with bioinformatics findings. For instance, mutagenesis studies are combined with sequence alignments and structural protein models to provide novel molecular views of protein function. Eventually, heterogeneous data such as gene expression profiles [7] and protein interaction networks need to be combined into time-dependent cellular models of disease mechanisms.
Lit.: 1. Albrecht M, Lengauer T, Schreiber S. Disease-associated variants in PYPAF1 and NOD2 result in similar alterations of conserved sequence. Bioinformatics 2003;19(17):2171-5. 2. Schreiber S, Rosenstiel P, Albrecht M, et al. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet 2005;6(5):376-88. 3. Albrecht M, Domingues FS, Schreiber S, Lengauer T. Structural localization of disease-associated sequence variations in the NACHT and LRR domains of PYPAF1 and NOD2. FEBS Lett 2003;554(3):520-8. 4. Stoll M, Corneliussen B, Costello CM, et al. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet 2004;36(5):476-80. 5. Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun 2005;327(3):679-87. 6. Valentonyte R, Hampe J, Huse K, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet 2005;37(4):357-64. 7. Costello CM, Mah N, Hasler R, et al. Dissection of the Inflammatory Bowel Disease Transcriptome Using Genome-Wide cDNA Microarrays. PLoS Med 2005;2(8):e199.1-17.


