Introduction
Breast cancer is a common neoplasm with a high mortality rate among woman with advanced stage disease. As is cur-rently believed for most cancers, one of the major factors contributing to cellular transformation and, ultimately, to inva-sive cancer is the cancer cell's ability to evade apoptosis. Although much has been learned about the molecules invol-ved in apoptotic signal transduction and its regulation, the identification and validation of molecular targets for apopto-sis-based cancer therapy still poses a major challenge. Stra-tegies addressing this challenge in the postgenomic era are genomewide screens for transformation related survival genes. The validity of such screens has been recently under-scored by a SAGE analysis of primary breast cancer material that led to the identification of survival factors with oncogenic potential (1).
In line with this, we have developed a genomewide screen for survival genes based on the cellular response to tumor necrosis factor alpha (TNF-alpha). As has been previously shown, TNF-alpha induces two intracellular protein com-plexes with opposite effects on cell survival (2). While one complex engages NF-#B and upregulates survival genes, the other initiates apoptosis by recruiting FADD and caspase-8. By specifically blocking the assembly of the apoptotic com-plex, we have identified several putative survival genes of which some might be useful as diagnostic markers and/or molecular targets for anti-breast cancer drug development.
Project Status
Experimental strategy
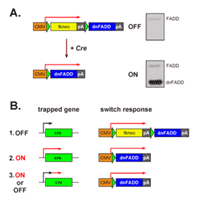
By combining gene trap mutagenesis with site specific re-combination (3), we have developed a strategy that enriches for survival genes induced by TNF-alpha in the human breast cancer cell line MCF-7. The strategy relies on a one way gene expression switch, which is activated by gene trap in-sertions into TNF-alpha inducible genes. As activation entails the expression of dominant negative FADD (dnFADD), a pro-tein that blocks the assembly of apoptotic complexes (4), the strategy enables the recovery of genes induced by the TNF-alpha dependent survival (NF-#B) complex (Figure 1).
Identification of TNF-alpha regulated genes
We have established a MCF-7 reporter cell line expressing the CMVlxtkneolxdnFADD switch cassette and used this cell line to construct an integration library consisting of 2x106 in-sertions of the retroviral gene trap vector U3Cre (5). From this library, we have isolated 66 cell lines with U3Cre inser-tions in TNF-alpha inducible genes. PCR amplification and sequencing of the genomic regions adjacent to the gene trap insertion sites (gene trap sequence tags; GTSTs) revealed that 28 GTSTs belonged to known genes, 12 corresponded to full length cDNAs with unknown function and 12 were part of expressed sequence tags (ESTs). Five GTSTs matched GeneScan predicted, transcribed regions and 9 correspon-ded to not yet annotated genomic regions. Among the known genes, some have been shown previously to be upregulated in various cancers, e.g. erbB3, ctnnd2, spata5 and S100A10. Moreover, the protein EMSY, encoded by one of the reco-vered genes, is directly linked to the development of sporadic breast cancers. As has been recently shown, the gene is amplified in more than 10% of sporadic breast and ovarian cancers whereby ist product EMSY inhibits the BRCA2 tumor suppressor protein (6). Accordingly, is believed that EMSY
overexpression is the functional equivalent to BRCA2 muta-tions typical for one inherited form of breast cancer. Collecti-vely, these results clearly underscore the validity of the gene trap approach.
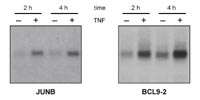
Validation of TNF-alpha induction of the trapped genes by Northern blotting revealed that the majority of them is regula-ted. Figure 2 shows two examples including JUNB, a known NF-#B target gene, and the human homolog of the mouse BCL9-2 gene, which thus far has not been associated with TNF-alpha signalling.
Identification of antisense transcripts
Closer inspection of the GTSTs revealed that the majority gene trap insertions occurred into the 5'-ends of genes, re-flecting the well known integration preference of retroviruses for promoter proximal regions. Interestingly, more than 40% of the proviral insertions occurred in opposite transcriptional orientation relative to the trapped gene, strongly suggesting the presence of naturally occurring antisense transcripts (NATs) initiated by Pol II promoters residing on the minus strand.
An in silico analysis of these antisense insertions using three different, publicly available promoter prediction programs, identified putative promoters on the minus strands in almost 50% of the cases. This indicates that the gene trapping stra-tegy employed here enables the recovery of non-coding, re-gulatory RNAs in addition to protein coding mRNAs that are regulated by a biological stimulus.
Outlook
Functional properties of candidate proteins
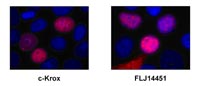
We have selected several TNF-alpha regulated candidate genes for more detailed functions analysis. Studies aimed at determining putative transforming and/or anti-apoptotic func-tions of the recovered proteins are in progress. Experiments involve protein overexpression exercises coupled with the analysis of intracellular protein distribution (Figure 3) as well as knock down approaches using RNAinterference (RNAi).
Analysis of antisense transcripts
Promoters predicted on the antisense strands of the trapped genes will be verified experimentally by cloning the respecti-ve genomic upstream sequences into luciferase reporter vec-tors. Promoter activity will be assessed in transient trans-fection asssays of MCF7 cells in the absence and presence of TNF-alpha. In addition, we will isolate the full length anti-sense transcripts and subject these to an in depth structural and functional analysis. Experiments will focus on their po-tential role in regulating gene expression. Finally, making sense of the antisense transcripts will greatly assist the on-going annotation of the human genome.
Lit.: 1. Porter et al. A neural survival factor is a candidate on-cogene in breast cancer. Proc Natl Acad Sci USA. 2003 Sep 16;100(19):10931-6. 2. Micheau and Tschopp. Induction of TNF receptor I-mediated apoptosis via two sequential signa-ling complexes. Cell. 2003 Jul 25;114(2):181-90. 3. Russ et al. Identification of genes induced by factor deprivation in hematopoietic cells undergoing apoptosis using gene-trap mutagenesis and site-specific recombination. Proc Natl Acad Sci USA. 1996 Dec 24;93(26):15279-84. 4. Hsu et al. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996 Jan 26;84(2):299-308. 5. Wempe et al. Gene trapping identifies transiently induced survival genes during pro-grammed cell death. Genome Biol. 2001 Jun 27;2(7): re-search 0023.1-0023.10. 6. Hughes-Davies et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003 Nov 26;115(5):523-35


