
Introduction
There is increasing evidence that the 3D organization of the genome has an essential impact on nuclear functions such as replication, gene expression and gene silencing. This is mirrored by the fact that nuclei of a defined cell type are generally characterized by a distinct morphology and chromatin texture, suggesting a cell type specific three-dimensional higher order chromatin architecture. These light microscopic observations, however, do not provide a detailed insight into the underlying mechanisms establishing and maintaining such a cell type specific nuclear architecture.
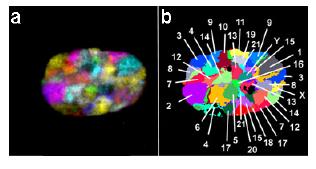
Fig 1: All chromosome territories, each in a different color, can be visualized simultaneously in intact interphase nuclei. (a) depicts an RGB image of the 24 differently labelled chromosomes (1-22, X, and Y), which was produced from deconvoluted mid-plane nuclear sections from a 3D stack by superposition of the seven color channels. The fluorochromes employed for probe labelling include diethylaminocoumarin (DEAC), Spectrum Green and the cyanine dyes Cy3, Cy3.5, Cy5, Cy5.5, and Cy7. (b) As in 24-color karyotyping, each chromosome can be identified by using a combination labelling scheme, in which each chromosome is labelled with a different set of fluorochromes [for details see Bolzer et al. (2005) or Speichar and Carter (2005)].
However, it is an unanswered question whether a premalignant state involves local or large-scale reorganization of the interphase genome. Under the assumption of a spatially sensitive difference between a low and high probability for a translocation, one might expect that already small-scale changes on the intranuclear chromatin arrangement can lead to profound cytogenetic events, such as structural or numerical rearrangements. These chromosomal changes, in turn, may result in dramatic changes in the gene expression profile of the respective cell, which may trigger the malignant transformation.
A comprehensive characterization of a cell type specific 3-D genome organization in normal and malignant cell types is still missing. A detailed characterization of the 3D-genome organization would provide the necessary basis to explore the potential impact of using higher order chromatin organization as a tool for the diagnosis and differential diagnosis of premalignant and malignant lesions. In contrast to array gene expression analyses, such topologic studies provide information on a single cell level and have therefore the potential to be of superior resolution. Regarding diagnostic pathology, digital imaging will increasingly replace conventional microscopes. Therefore it is conceivable, that 3D analyses become part of the routine diagnostic algorithm of cell and tissue samples. Possible diagnostic scenarios could include cytological evaluation of buccal smears (individuals with high risks for head and neck cancer), urine sediment (individuals at risk for bladder cancer, treatment monitoring) or Pap-smear (cervix cancer). Regarding tissue sections, this technique can be applied to the morphological spectrum of colonic mucosa, ranging from normal and hyperplastic epithelium to various types of adenomas and carcinomas, and as another example can be integrated into the differential diagnosis of ovarian tumors which are characterized by large variation in phenotype and biological behavior.
Project Status
Multicolor Interphase FISH
The project partners Michael Speicher and Thomas Cremer have recently shown that all 24 chromosome territories can simulatenously be visualized in intact human fibroblasts (Fig. 1). This represents an extraordinary achievement and allows a detailed analysis of higher order chromatin organization in human nuclei. These analyses indicate that the arrangement of chromosome territories may be different in fibroblasts as compared to lymphocytes. In the former cell type the size of chromosomes seems to be an important determinant of chromosome position, while in the latter cell type the gene density of the respective chromosomes has an impact whether a chromosome will be in the center or in the periphery of a nucleus. Beside the detailed analysis of single cell suspensions there will be an especial emphasize on biological samples, such as tissue sections. To this end we have extended our previous protocol for the simultaneous hybridization of up to 7 differently labeled chromosome centromere specific probes on thick tissue sections (>20 µm). Our imaging and software analyses tools provide a unique mean to investigate the chromosomal composition within any tissue source. Figure 2 illustrates such an example, in which chromosome numbers were determined in a nucleus of a colorectal adenoma.
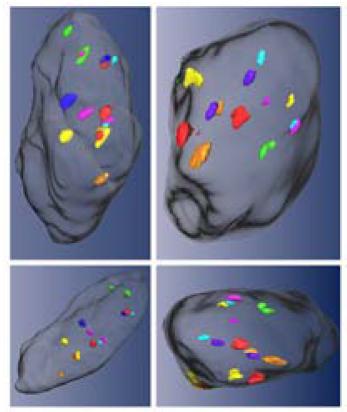
Fig 2: Simultaneous visualization of seven different centromeres probes (chromosome 3: green; 4: light blue; 7: orange; 8: pink; 17: yellow; 18: blue; 20: red) in a nucleus of a colon adenoma. The same nucleus is depicted from several different angles.
Furthermore, our method development will be used for the acquisition of tissue samples (normal, premalignant states, malignant states), such as cytological smears and fluids, histological specimens of normal and neoplastic colonic mucosa as well as non-neoplastic and neoplastic ovarian tissue. In addition, the various technologies will be applied to primary cell cultures of normal and neoplastic cells, e.g. ovarian surface epithelium and tumors.
As already mentioned, we are establishing improved probe sets for a number of different applications. To include functional information, the probe set design will be based on expression analyses data of the respective tissue. This will ensure that the probe set will not consist of randomly chosen genomic regions, but of regions for which an important functional role has been established. For these regions position effects under certain pathologic conditions should be most likely. Thus, we will screen published expression analysis data, identify BACclones, which cover relevant genes, generate multicolor BAC-probe sets (at least 7 different BAC-clones) and use them for the interphase screen. Establishment of the 3D-topology in normal cells will be followed by a comparison with cells in defined disease states.
Microscopy development
The elucidation of a 3D-topology of different cell types can only be achieved by a high-resolution analysis of a large number of nuclei. We anticipate that the analysis of at least 1.000 nuclei/cell type is needed to determine the 3Dorganization of the genome. A similar number of nuclei will have to be evaluated for defined disease stages. This can be achieved by using an automated high-throughputplatform (iMICTM) for the multicolor 3D-evaluation. It uses spinning-disk-technology in combination with a noncoherent light-source, which is directly coupled to the scanning system through a patented quartz light-guide. It allows the excitation with a multitude of freely selectable wavelengths between 340 and 760 nm and with adjustable bandwidth. Two emission-colors are collected simultaneously on a single chip of a CCD-camera. It is planned to employ recent on-chip-amplification technology which allows single photon counting, and to combine this feature with back-thinned CCDs which provide quantum efficiencies in excess of 90%.
Within the framework of this NGFN project we have teamed up with our industrial partner TILL-Photonics. The aforementioned iMIC platform has been developed by this company. Another task will be the adaptation of appropriate software tools for 3D-signal identification. The latter will employ novel deconvolution algorithms, which can be specifically optimized for the geometry of the fluorescent target, for the distance measurements and for the automated evaluation.
Outlook
Our ongoing efforts will yield the following deliverables: Establishment of 3D genome organization for defined tissue types and detailed comparison of the topology in normal tissue vs. disease states (e.g. premalignant state, malignant state). The development of optimized probe sets, which may be also of relevance for diagnostic applications. Development of a high-throughput, high content platform for the automated screening of large cell numbers after hybridization with multiple-FISH probes. The aim is to develop a microscope combining the spatial and spectral resolution of the best classical confocal microscopes with the versatility of a CCD-based multicolor system. It will employ non-coherent light-sources of unprecedented brightness, thus not only making a great number of excitation wavelengths available, and it will permit a level of throughput, a degree of automation and a long-term stability and robustness not available with laser-scanning approaches. Both, the establishment of a disease associated topology and a high throughput microscopy platform may evolve to new, powerful diagnostic tools.
Lit.: 1. Cremer M, et al. Inheritence of gene-density related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol 2003 162:809-20. 2. Bolzer A, et al. Three-dimensional maps of all chromosome positions indicate a probabilistic order in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 2005 3:e157. 3. Speicher MR and Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005 6:782-92. 4. Mayr C et al. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2005 Sep 22; [Epub ahead of print].


