Introduction
Infection with human cytomegalovirus (HCMV), a #-herpesvirus, is a major risk factor for immunoincompetent and immunocompromised patients (opportunistic pathogen). Infection of mice with murine CMV (MCMV) represents the rodent model. Both viruses compromise the innate and adaptive immune response by numerous immunomodulatory viral gene functions. Viral proteins affect MHC class I and II antigen presentation, interfere with the interferon response and control apoptosis (ref 1). A recent finding is that CMV genes affect innate NK cell response (ref. 2) and the inflammatory response by viral components that act on Toll-like receptors (TLR) (ref 3). Here we wish to define the role of viral genes that affect the innate immune response in vitro and in vivo.
NK cells dominate the first phase of CMV infection (ref 2). We and others have identified three viral genes of MCMV that interfere with NK cell activation. m144 encodes an MHC class I homologue and inhibits NK response by a so far unknown mechanism (ref 4). m157 turned out to directly trigger the activatory NK cell receptor Ly49H in C57BL/6 mice and thereby explains the resistance of this mouse strain to MCMV infection (ref 5). The third gene, m152, encodes a protein that prevents surface expression of Rae-1, a ligand for the activatory NK cell receptor NKG2D (ref 6). Using a set of large deletion mutants we confirmed the existence of at least a fourth NK regulator among ten genes. We wish to identify and characterize this additional NK regulator in vivo.
Dendritic cells (DC) participate in both innate and adaptive immunity and play a crucial role in the outcome of an immune response to microbial pathogens (ref 7). Therefore, modulating DC functions by targeting DC gene expression is an attractive target for CMV to evade from immune attack. In the past, analysis of viral gene function was mainly carried out in fibroblasts although this cell type is only of little relevance for CMV infection in vivo. We wish to use microarray analysis to determine which cellular genes are differently expressed during DC by MCMV infection. Subsequently the viral genes responsible for these effects will be identified by screening a library of deletion mutants in vitro.
The major problem of using microarray analysis to investigate virus-host cell interaction in vivo is the high background signal of uninfected cells and the arising general inflammatory response leading to numerous changes of gene expression in a variety of cell types. We want to eliminate these background signals by selectively labeling and isolating RNA from infected cells using the activity of uracil phosophoribosyl transferase from toxoplasma gondii (ref 8).
Uracil phosphoribosyltransferase defines the pyrimidine salvage pathway in many organisms. It is utilized by prokaryotes as well as eukaryotes, however it is not present in mammals. Besides its normal substrate - uracil – uracil phosphoribosyltransferase from toxoplasma gondii (Tg-UPRT) is able to utilize 4’-thiouracil (4TU) as an additional substrate. Incubating cells expressing Tg-UPRT with 4TU leads to the formation of 4-thiouridine monophosphate. After two additional phosphorylation events catalysed by cellular kinases the resulting 4-thiouridine triphosphate is incorporated into newly synthesized RNA. As thiol groups are usually not present in mammalian RNA this thiol-labeled RNA can be specifically tagged with a thiol-selective, reversible biotinylation reagent (Biotin-HPDP) which leads to the formation of a disulfide bond between 4TU and Biotin-HPDP. Streptavidin coated magnetic beads can be used to isolate biotinylated RNA from total RNA. 4TU-labeled RNA is eluted from the beads using a reducing agent (DTT) which cleaves the disulfide bond thereby leaving the biotin to the streptavidin and restoring the original RNA. This RNA can now be used for microarray analysis.
We want to use this method to selectively label newly synthesized RNA in virally infected cells by introducing Tg-UPRT into the genome of murine cytomegalovirus (MCMV). This will allow us to selectively purify cellular and viral RNA out of virally infected cells on a background of uninfected cells. With this technique we aim at studying virus-host interactions on gene expression and RNA decay in vitro and in vivo.
To evaluate the role of new immunomodulatory candidate genes in MCMV infection of DC in the physiological context in vivo we have established a new tool – namely conditional mutagenesis using cre-recombinase (ref 9).
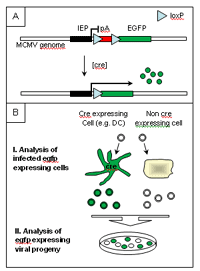
We have constructed a virus that expresses egfp only after cre-recombinase mediated sequence excision (Fig 1A). Infecting mice expressing cre-recombinase under a DC specific promotor with this virus should lead to activation of egfp only in infected DCs. This should allow to determine the contribution of DC in MCMV infection by analyzing the labeled virus progeny generated by DC (Fig 1B). Introduction of mutations into the MCMV genome and depletion protocols or genetic deletion of host innate immune functions (e.g. TLR) will provide novel in vivo readouts to evaluate viral functions determined by microarray screening and other means.
Results/Project Status
We were successful in identifying and characterizing an additional NK modulatory gene in the genome of MCMV. m145 encodes a viral protein that abolishes the surface expression of murine UL-16-binding protein-like transcript (MULT-1), a cellular ligand for the activatory NK cell receptor NKG2D. We could show that m145 compromises virus control by NK cells in spleen and lungs of infected mice.
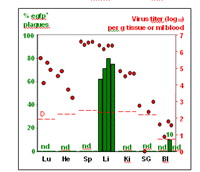
We could also show that conditional mutagenesis can be used efficiently to modify viral genomes cell type specifically in vivo which allowed us to evaluate the role of certain cell types during an MCMV infection in the organ context. We infected mice expressing cre recombinase under control of the hepatocyte specific albumin promoter (Alb-cre) with a recombinant MCMV containing a cre activatable egfp reporter gene. Liver was the only solid organ of infected mice were egfp expressing infectious virus could be detected proving high specificity and efficiency of cre mediated recombination excluding ectopic activation of egfp (Fig 2).
Regarding the progress on selectively labeling and isolating RNA of infected cells using Tg-UPRT we have now inserted the UPRT gene into the MCMV genome using BAC-technology and reconstituted the virus. 24 hours after infection of murine M2-10B cells with this virus and after further three hours of incubation with 4TU we were able to selectively purify about 10% of total RNA, i.e. RNA synthesized during the incubation with 4TU. No RNA could be purified from mock-infected cells.
Outlook
As we have hints that even a fifth NK regulator is encoded by the genome of MCMV we are currently trying to identify and characterize it. Once these five NK regulators are known a mutant MCMV will be generated lacking all five of them. With this mutant the impact of MCMV on NK cells control can be studied.
To further evaluate whether Tg-UPRT is a useful tool in selectively isolating RNA from infected cells we are currently preparing virus-stocks to start the first in vivo experiments. Once the method has been shown to yield sufficient amounts of labeled RNA from in vivo experiments we want to perform microarray analyses and investigate the effects of virus-host interaction on gene expression of cellular and viral genes in different organs and cell types. In the following we want to compare different virus mutants to identify the responsible viral genes.
After we have proven that conditional mutagenesis is a highly efficient and specific tool to analyse tropism of MCMV to hepatocytes we wish to test this assay system with mice expressing cre in dendritic cells (DC) to be able to evaluate the role of this cell type during MCMV infection in vivo. Currently transgenic mice expressing cre recombinase under the control of the dendritic cell specific CD11c promoter are evaluated in their potential to activate the egfp reporter in the MCMV genome and to produce viral progeny. The conditional expression of a reporter (egfp) by the virus, expressed once it has passaged through a target cell important for innate immunity, provides a unique tool. We will explore wild type MCMV and mutants found by microarray screening with regard to several aspects leading to DC infection. Once the virus has infected DC the effects of innate immune control of virus replication in DC is open to analysis.
Lit.: 1. Alcami A and Koszinowski UH. Viral mechanisms of immune evasion. Immunology Today 2000 Sept. 447-455. 2. Lopez-Botet M. et al. Natural killer cell receptors for major histocompatibility complex class I and related molecules in cytomegalovirus infection. Tissue Antigens 2004 63; 195-203. 3. Tabeta K. et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. PNAS March 2004; 3516-3521. 4. Farrell HE et al. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature 1997 April 3; 510-514. 5. Arase H et al. Direct recognition of Cytomegalovirus by Activating and Inhibitory NK cell Receptors. Science 2002 May; 1323-1326. 6. Lodoen M. et al. NKG2D-mediated Natural Killer Cell Protection Against Cytomegalovirus Is Impaired by Viral gp40 Modulation of Retinoid Acid Early Inducible 1 Gene Molecules. J. Exp. Med 2003 May 19 ; 1245-1253. 7. Rinaldo CR Jr, Piazza P. Virus infection of dendritic cells: portal for host invasion and host defense. Trends Microbiol. 2004 Jul;12(7):337-45. 8. Cleary MD et al. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nature Biotechnology. 2005 Feb 23; 232-37. 9. Branda CS and Dymecki SM. Talking about a Revolution: The Impact of Site-Specific Recombinases on Genetic Analyses in Mice. Developmental Cell 2004 Jan; 7-28.


