Introduction
A considerable proportion of “idiopathic” cardiomyopathies (~30%) is due to genetic causes1 and several disease genes for hypertrophic (HCM) and dilated cardiomyopathy (DCM) have been identified in affected patients2. Most of these genes encode for sarcomeric or cytoskeletal proteins, but the precise mechanisms and signaling events that translate these mutations into a cardiomyopathic phenotype in affected patients remains poorly understood.
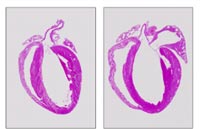
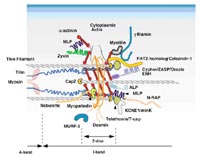
The sarcomeric z-disc, a structure tradionally believed to simply crosslink myofilaments, is emerging as a “hot spot” for cardiomyopathy disease genes, including muscle-LIM protein (MLP), cypher/ZASP and T-Cap/telethonin. Moreover, it provides an important link between structural proteins and cardiomyocyte signaling3. We have recently identified a novel protein family of muscle-specific z-disc proteins, termed calsarcins4,5, which directly interact with cypher/ZASP and T-Cap/telethonin. Calsarcins also bind the phosphatase calcineurin, which plays a critical role in the transduction of calcium signals leading to cardiomyopathy6,7. We have now generated calsarcin-deficient mice, which reveal a cardiomyopathic phenotype in response to biomechanical stress8 (Fig 1).
Results/Project Status and Outlook
Our first specific aim was to perform yeast-two hybrid assays with z-disc associated proteins either implicated in human DCM or associated with cardiomyopathy in animal models, in order to identify novel binding partners. Several of these screens have been completed successfully and have revealed novel binding partners for z-disc proteins, including calsarcin, MLP, affixin and gamma-filamin.
In a complementary approach, we plan to characterize novel genes with a potential role in disease pathways. During NGFN1 we found a set of >100 novel/uncharacterized ESTs to be differentially regulated in human DCM and/or experimental cardiomyopathies, such as calcineurin-trangenic mice6, calsarcin-1 knock-out mice8, and Troponin T transgenic rats10. The second specific aim was to systematically analyze these clones in vitro utilizing semi-automated assays (in close cooperation with SMP Cell). These experiments allow to simultaneously examine a large number of candidates for their basic properties, including effects on cell growth, cell death and selected signal transduction pathways as well as the determination of their subcellular localization in muscle cells11. The third specific aim will be to characterize promising ESTs (e.g. with a muscle-specific expression pattern) in detail by yeast-two-hybrid screens with cardiac cDNA libraries, siRNA-mediated “knock-down” in neonatal cardiomyocytes (including subsequent microarray analyses) and the generation of loss of function mutations in zebrafish. A subset of the most interesting molecules (e.g. those that are associated with cardiomyopathy in zebrafish) will also be studied in mouse models, either by transgenesis or generation of “knock-out”-models. Finally, the fourth specific aim will be to test novel genes that reveal a cardiomyopathic phenotype in animals for mutations in the DCM patient cohort.
Lit: 1. Grünig E, Tasman JA, Kücherer H, Franz WM, Kübler W, Katus HA. Frequency and phenotypes of familial dilated cardiomyopathy. J. Am. Coll. Cardiol. (1998) 31, 186-194. 2. Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell. Dev. Biol. (2002) 18, 637-706. 3. Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell (2002) 111, 943-55. 4. Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc. Natl. Acad. Sci. USA (2000) 97, 14632-14637. 5. Frey N, Olson EN. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. (2002) 277, 13998-14004. 6. Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215-28. 7. Frey N, McKinsey TA, Olson EN. Decoding calcium signals involved in cardiac growth and function. Nat. Med. (2000) 6, 1221-7. 8. Frey N, Barrientos T, John Shelton, Derk Frank, Hartmut Rütten, B. Rothermel, James A. Richardson, Hugo A. Katus, Joseph A. Hill, Olson EN. Mice lacking calsarcin-1 are sensitized to calcineurin signalling and display accelerated cardiomyopathy in response to biomechanical stress. Nat. Med. (2004) 10, 1336-43. 9. Frey N, Olson EN. Cardiac hypertrophy: The Good, the Bad, and the Ugly. Annu. Rev. Physiol. (2003) 65, 45-79. 10. Frey N, Franz WM, Gloeckner K, Degenhardt M, Müller M, Müller O, Merz H, Katus HA. Transgenic rat hearts expressing a human cardiac troponin T deletion reveal diastolic dysfunction and ventricular arrhythmias. Cardiovasc. Res. (2000) 47, 254-264. 11. Wiemann S, Bechtel S, Bannasch D, Pepperkok R, Poustka A and The German cDNA Network. The German cDNA Network: cDNAs, functional genomics and proteomics. Journal of Structural and Functional Genomics (2003) 4, 87-96.


