Introduction
Myocardial infarction (MI) is both multifactorial in origin and phenotypically diverse. Multiple etiologic components and multiple morphological features of CAD make it likely that different causes lead to different disease patterns. Thus, in addition to differentiate factors leading to CAD, it is crucial to precisely specify distinct features of coronary morphology. Accordingly, we hypothesize intermediate phenotypes re-flecting certain aspects of CAD facilitate the identification of distinct gene-phenotype relationships. Particularly, the phe-notypic characterization of the coronary morphology may enable to specifically investigate whether distinct CAD pat-terns reflect distinct genetic causes. The importance of such substratification has recently been documented in the identi-fication of the first stroke-related gene.1
Project Status
Studies in humans – heritability of coronary phenotypes
We retrospectively studied the coronary angiograms of 882 siblings with CAD from 401 families (Figure 1). These families were ascertained through index patients defined by MI before the age of 60 years and at least one sibling with MI or coronary revascularization procedures. Heritability calcula-tions were performed using variance component analysis. Additionally, recurrence risks to siblings were analyzed.4
Traditional cardiovascular risk factors and age at the first coronary event displayed significant heritable components. After adjustment for age and gender, significant heritabilities were identified for proximal stenoses, in particular left main disease (h2=0.49#0.12; p=0.01), coronary calcification (h2=0.51#0.17; p=0.001), and ectatic coronary lesions (h2=0.52#0.07; p=0.001) (Figure 2).
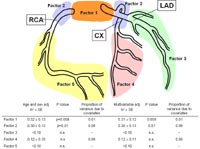
In contrast, no heritability was found for distal disease (h2=0.05#0.19; n.s.), the pattern of coronary arterial blood supply or the number of diseased vessels (Fig. 2).
To confirm the heritability measures we analysed a second set of 1,500 coronary angiograms from MI families and tested for heritability of selected coronary phenotypes.
The analyses are ongoing; but preliminary results confirm the heritability measures in this second sample.
Animal studies – unravelling the Dyscalc1 locus
Dyscalc1 was mapped to a chromosomal region on mouse proximal chromosome 7. Since, Dyscalc1 was intensively investigated. We localized Dyscalc1 to a 10 cM chromoso-mal segment between D7Mit227 and D7Mit230 microsatellite markers. We confirmed the contribution of Dyscalc1 to dys-trophic cardiac calcification (DCC) using a congenic line that carry the DCC-susceptible allele on a resistant genetic back-ground.5 We demonstrated that Dyscalc1 contribute not only to cardiac calcification but also to aorta calcification.6 Fur-thermore, using bone marrow transplantation, we showed that calcium phosphate deposit was initiated in DCC-susceptible strains but influenced by infiltrating bone marrow cells.7
The aim of the ongoing project is to identify, among the 160 genes located in Dyscalc1 critical region, putative candidate genes representing the so far unknown Dyscalc1 gene(s).
Differential gene expression
On the basis of differential gene expression patterns, which were determined in various tissues from resistant C57BL/6 and susceptible C3H/He mice, we identified putative candi-date genes in the Dyscalc1 critical region. Specific primer pairs for each gene of the 160 genes located in Dyscalc1 interval were designed and polymerase chain reaction (PCR) has been performed for detecting the gene expression in different organ tissue. Genes that were expressed in myo-cardial tissue have been further investigated using Real-time RT-PCR (Figure 3).

Sequence analyses of putative candidate genes are ongoing and experiments are underway to further examine the bio-logical functions of genetic variants in these candidate genes in mice and humans.
In-silico mapping of Dyscalc1 critical region
In a complementary approach we further tried to narrow down the Dyscalc1 critical region using “in silico”-mapping.8
Analyzing recombination among eight laboratory strains within the Dyscalc1 segment further narrowed down this region to a 1-Mb segment within an “in silico” QTL peak with a significant R value of 1.8 (Figure 4). These results demon-strate that “in silico”-mapping is a powerful tool that could be used to confirm fine mapping results of classical QTL analysis.

Human Studies – Identification of genetic variants in candidate genes and association studies
Until now, we sequenced five candidate genes (5´UTR, 3´UTR, promoter, and coding exons) within the Dyscalc1 critical region in a subgroup of 48 patients with different degrees of coronary calcification.
The frequency of variants leading to amino acid substitutions will be further determined in larger sets of patients with different coronary phenotypes and controls. Our aim is to identify genetic variants associated with specific coronary phenotypes.
Lit.: 1. Gretarsdottir S et al. (2003) The gene encoding phos-phodiesterase 4D confers risk of ischemic stroke. Nat Genet. 35:131-8. 2. Ivandic BT et al. (1996) A locus on chromosome 7 determines myocardial cell necrosis and calcification (dys-trophic cardiac calcinosis) in mice. Proc Natl Acad Sci U S A. 93:5483-8. 3. Ivandic BT et al. (2001) New Dyscalc loci for myocardial cell necrosis and calcification (dystrophic cardiac calcinosis) in mice. Physiol. Genomics 6:137-44. 4. Fischer M. et al. (2005) Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation 111, 855-862. 5. Aherrahrou et al. (2004) A locus on chromosome 7 determines dramatic up-regulation of osteopontin in dystrophic cardiac calcification in mice. Am J Pathol. 164:1379-87. 6. Kazmarek et al. (2005) Reduced cardiac calcinosis in DCC-susceptible congenic mice recon-stituted with DCC-resistant bone marrow cells. Poster pres-entation; European Society of Cardiology, Stockholm, Swe-den. 7. Doehring et al. (2005) Dyscalc1 determines aortic calcification in mice. Poster presentation; European Society of Cardiology, Stockholm, Sweden. 8. Grupe A et al. (2001) In silico mapping of complex disease-related traits in mice Science. 292(5523): 1915-8.


