Introduction
Proteome analysis is a powerful methodology to investigate protein expression in tissues involved in diseases not linked to particular genetic defects such as Alzheimer’s and Parkin-son’s disease (AD and PD). Direct measurement of protein expression is essential for the analysis of these diseases, since posttranscriptional and -translational control such as splicing of mRNA, posttranslational modification of proteins (e.g. phosphorylation, ubiquitinylation, oxidation etc.) cause a non-predictive correlation between mRNA and protein levels. Differential proteome analyses comparing diseased and non-diseased brain tissue will result in the identification of target proteins associated with AD and PD. In collaboration with different partners of the SMP Proteomics consortium, the KG Neuro, and from international collaboration partners in the HUPO HBPP brain material of different mouse models is analyzed using the 2D-DIGETM technology (Fluorescence Difference Gel Electrophoresis system, GE Healthcare) [1]. Together with Prof. Dr. Auburger, Molecular Neurogenetics, JW Goethe University of Frankfurt two different PD mouse models (#-synuclein transgenic and knock-out model) are under investigation. Prof. Auburger and colleagues devel-oped a transgenic mouse model with mutated #-synuclein A53T [2]. This mutation was found to be the cause of auto-somal dominant early onset PD in a small group of Meditter-anean families and is identified as a major constituent of the “Lewy body” protein aggregates. In differential proteome studies comparing protein patterns of diseased and non-diseased mice the proteomes of different distinct develop-mental age stages are compared within this subproject. The results are compared with those derived from analysis of the #-synuclein knock-out model potentially giving more insight the role of #-synuclein in PD.
Another focus is on the analysis of phosphorylated, ubiquiti-nated and oxidized proteins in an AD UBB+1 transgenic model in close collaboration with Prof. Dr. van Leeuwen, Netherlands Institute for Brain Research (NIBR), Amsterdam. At the NIBR a frameshift mutant of ubiquitin (UBB+1) was identified to be a reporter for a dysfunction of the proteasome contributing to the neuropathogenesis of AD or other tau-opathies. The presence of ubiquitin and ubiquitinated pro-teins in the proteasome indicate a dysfunction of the protea-some leading to inhibited protein degradation and increased aggregation. Besides ubiquitination phosphorylation and dephosphorylation of different proteins (tau, presenilin by kinases such as CDL-5 und GSK3#) play a major role in AD [3, 4, 5]. To further understand the downstream events asso-ciated with the appearance of UBB+1, the NIBR has gener-ated novel transgenic mouse lines expressing UBB+1 at high levels in the brain and in a region specific. The obtained data will serve as basis for further differential examination of post-mortem brain material of AD and PD patients in comparison to control brain tissue.
Today, most differential proteome studies are performed using two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) [6, 7, 8] as the state-of-the-art technique for the separation of highly complex protein mixtures. This tech-nique indeed is subject to several limitations (linearity is limited to 2-3 orders of magnitude, high molecular weight, very acidic/basic and membrane proteins are underrepre-sented). To overcome these restrictions new technologies have to be developed, e.g. LC-based methods. In our sub-project a new high sensitive multi-dimensional (MD) tech-nique for the separation of intact proteins in a high dynamic range is developed. For this purpose the DIGETM technology is adapted for quantitative LC-based proteome analyses.
Project Status
Differential proteome analyses of age-related mouse models - The #-synuclein PD model and UBB+1 AD model
In both studies the protein patterns of brain tissue of dis-eased and non-diseased control mice are compared result-ing in the identification of potential biomarkers for AD and PD. For the separation and detection of differential proteins the 2D-DIGETM system (minimal labelling technique) is used. Proteins of interest are excised from the gels and digested using different proteases such as trypsin. Resulting peptides are analysed by mass spectrometry. The obtained data are automatically interpreted using different search algorithms (SequestTM, Mascot) and software tools (ProteinscapeTM, DTASelectTM).
Differential proteome analysis of PD mouse models
In our differential proteome study, striatum, brain stem and cerebellum from A53T #-synuclein transgenic mice and FVB/N (control) mice are dissected at different stages of age. The differences in the brain proteome between these mouse lines will be determined to elucidate the role of #-synuclein at different stages of PD.

First experiments have shown that our preparation protocol extracts sufficient proteins out of small tissues like striatum or brain stem to run several 2D-gels. Three different age stages (6, 12 and 22 months of age) of each model – the #-synuclein transgenic and the knock-out mouse model – have to be differentially compared with the respective controls. To date the differential analysis of the brains of the #-synuclein transgenic mouse model (stages 6 & 22 months) were com-pleted. In total 22 differential proteins were detected and 12 of these could be identified up to now. The stages of 12 months will follow and data will be validated soon. In the next step the knock-out mice of the stages 6 & 22 months will be analysed as well as cerebella of transgenic, knock –out and control mice.
Differential proteome analysis of an AD mouse model
Our specific aim is to elucidate a number of downstream targets in transgenic mice with UBB+1-mediated UPS inhibi-tion, such as phosphorylated, oxidised and ubiquitinated proteins. For this purpose protein patterns of cortices from the UBB+1 transgenic and control mice are differentially analysed at different stages of age (3, 9, 15 months). Post-translationally modified proteins are detected after 2D-gel electrophoresis using different methods, e.g. antibodies and a phosphoprotein specific fluorescence stain (ProQDiamond, Invitrogen).

Different proteins are identified so far using a hybrid triple quadrupole/linear ion trap mass spectrometer – the 4000 Q Trap®. The first results demonstrate that all techniques provide a good accession to the differential phosphopro-teome analysis of the UBB+1 transgenic mice: Several dif-ferentially phosphorylated proteins are already detected after ProQDiamond staining and mass spectrometric analysis of one spot successfully resulted in localisation of a phosphory-lation site in HSP 90b.
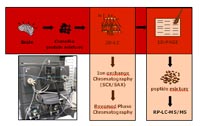
Quantitative LC-based protein analysis
An alternative method overcoming the limitations of 2D-PAGE provides multidimensional chromatography. There-fore, proteins are mostly digested and resulting peptides are separated by MD-HPLC; e.g. MudPIT is one of these meth-ods. Nevertheless, the complexity of the sample is enor-mously increased analysing peptide in contrast to protein mixtures and the dynamic range is only about 102 hampering the detection of low abundant proteins. For that reasons there is high demand to separate complex mixtures on the basis of intact proteins.
In our project, we evaluate and establish a multidimensional chromatography-based protein separation method enabling comprehensive protein detection in a dynamic range of about 108. The combination of this technique with differential quan-titative protein analysis will allow the identification of even low abundant target proteins associated with e.g. neurode-generative diseases. Proteins from brain lysates are first fractionated by strong anion exchange (SAX) or strong cation exchange (SCX), respectively. We will start with about 100 mg of protein for the separation in the first dimension (SAX or SCX, respectively). Resulting fractions are collected and transferred to a second LC-based protein separation step (RP-HPLC). In the third step the proteins are separated by 1D-SDS-PAGE. Quantification will be first done optically by fluorescence detection. These experiments will be repeated 3 to 5 times to prove the reproducibility of the quantitative results. Thereafter, differentially detected protein bands will be proteolytically digested and the resulting peptide mixtures will be analyzed using nano-rpLC-MS/MS.
In first experiments we established a separation technique including SAX chromatography in the first and RP chroma-tography in the second dimension using pig brain proteins lysates. Fractionated proteins are separated using large high resolution SDS-PAGE (20 x 20 cm). Reiterations of the experiments exhibit a high reproducibility of both chroma-tographic (first and second dimension) and gel electropho-retic level, emphasizing the applicability of the designed prefractionation technique.
Outlook
Results obtained from the mouse model studies will be validated in human material, i.e. preparations of selected human brain regions of donors who suffered from AD or PD, or of neurologically healthy control donors. We hope to link changes in AD and PD patients to the molecular pathways disturbed by the transgenesis in the analysed mouse model, and to better understand these multifactorial diseases. New avenues for therapy will be discovered: a number of proteins, e.g. modified and trapped in aggregates at different age stages will be known, and mouse models based on this knowledge will have shown their individual contribution to neurodegeneration.
After optimization of the multidimensional HPLC with sam-ples of pig brain protein lysates the method is usable for the analysis of high complex protein mixtures. The next step is the implementation of the DIGETM technique to this chroma-tographic approach allowing differential quantitative analysis. These new high-sensitivity methodic developments will explicitly advance the performance of current proteomic analyses. The aim is the identification of differential low abundant proteins serving as target molecules for the diag-nosis and therapy of diseases - such as PD and AD - not detectable by other analysis techniques.
Lit.: 1. Alban A et al. A novel experimental design for com-parative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003 Jan 3;1:36-44. 2. Gispert S et al, Transgenic mice expressing mutant A53T human alpha-synuclein show neuronal dysfunction in the absence of aggregate formation. Mol Cell Neurosci. 2003 Oct 24;2:419-29. 3. Yoshimura Y et al., Phosphorylation of tau protein to sites found in Alzheimer's disease brain is catalyzed by Ca2+/calmodulin-dependent protein kinase II as demon-strated tandem mass spectrometry. Neurosci Lett. 2003. Dec 26;353(3):185-8. 4. Fluhrer R et al., Phosphorylation of presenilin 1 at the caspase recognition site regulates its proteolytic processing and the progression of apoptosis. J Biol Chem. 2004. Jan 16;279(3):1585-93. 5. Lu KP et al Proline-directed phosphorylation and isomerization in mitotic regulation and in Alzheimer's Disease. Bioessays. 2003. Feb;25(2):174-81. 6. Klose J., Large-gel 2-D elec-trophoresis. Methods Mol Biol. 1999; 112:147-72. 7. O`Farrell PH, High resolution two-dimensional electrophore-sis of proteins.J Biol Chem. 1975. May 25; 250 (10):4007-21. 8. Görg A et al., The current state of two-dimensional elec-trophoresis with immobilized pH gradients. Electrophoresis. 2000. Apr;21(6):1037-53.


