
Introduction
The analysis of gene expression patterns has been demonstrated to be one of the most powerful and direct ways to systematically generate data for functional studies, including the understanding of gene function in development and differentiation. Within less than 10 years, gene expression profiling with microarrays has become a key technology in biomedical research. Gene expression analyses of large numbers of patient samples have been providing significant advances to the classification of diseases and the identification of target genes relevant to human disease for therapeutic, diagnostic, and prognostic applications. In addition, microarray data have made important contributions to our understanding of the regulation of biological networks and gene function.
This knowledge is fundamental to the elucidation of the entire network of changes causing and caused by a disease, and an absolute requirement for the knowledge-driven medicine of the future.
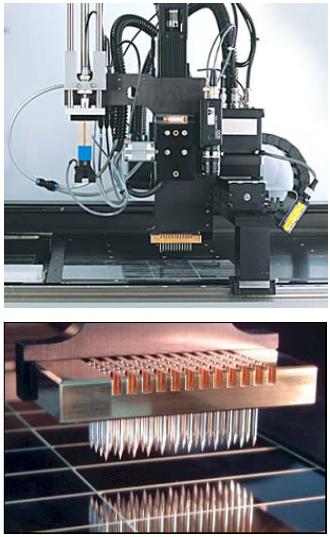
The gene expression profiling platform at the DKFZ is focusing on the development of technologies for high throughput gene expression analyses and their application to fight human diseases. The common goal of all activities is the elucidation of disease pathomechanisms. The core unit performs collaborative projects with the Disease-oriented Genome Networks (KGs) and other Systematic Methodological Platforms (SMPs) as well as separate research projects. The strengths of the platform have been documented in numerous scientific collaborations and publications.
On the platform, the experimental protocols for standardized array production (handling of large clone collections, PCR amplification of clone inserts by robotic pipetting systems, spotting of microarrays in a controlled environment; Figure 1) sample treatment (isolation, amplification and labeling), internal controls (negative controls, spiking with Arabidopsis thaliana reference RNA samples), as well as automated hybridization routines are established. From each microarray batch, 10% of the microarrays are routinely scanned after spotting. All slides are checked for remaining background before hybridization. DNA on two selected microarrays from each batch is stained with fluorescent DNA-binding dyes (Syto 61) to control for the presence of target.
All relevant protocols are published and accessible for the scientific community on the NGFN website (www.ngfn.de). We routinely apply human and mouse whole genome microarrays for microarray analyses in the NGFN.
Specifically, our arrays include:
(36,000 fully resequenced and bioinformatically characterized human genes/ESTs; RZPD Unigene 3.1). This clone set is currentlythe most comprehensive cDNA resource for microarray construction. It includes the complete collection of the
currently 20,000 ENSEMBL-annotated cDNA clones and approx. 16,000 ESTs. It has been distributed to multiple users, among them partners in the EU.
Apart from microarray analyses, the core unit is increasingly working on the validation of gene expression data (e.g. by quantitative RT-PCR). New technologies (e. g. RNA and signal amplification) are constantly developed and implemented into the laboratory routines. Most importantly, the integration of strong bioinformatic know-how working i
immediate collaboration with the experimental projects has proven to be highly successful in the past and is being continued in SMP-RNA.
Many collaborative projects with clinical partners have led to a wealth of information on gene expression in various tumors and other diseases. An example is mentioned below:
Diagnostic gene expression profiles in kidney cancer
Adult renal cell carcinoma (RCC) is one of the ten most common human malignancies in developed countries. Its global incidence has been increasing continuously during the past 30 years. RCC is divided into clear cell (ccRCC; 80 % of all cases), papillary (pRCC, 10 %), chromophobe (chRCC; 5 %), and several other rare types. Although the histopathological diagnosis of kidney cancer is well established in the clinical routine, the molecular basis for the distinction of RCC types is poorly understood. We constructed RCC specific cDNA microarrays and hybridized these with labeled cDNA derived from tumor samples of the three major RCC types. By using the microarray data of 35 RCC samples, we identified an 18-gene-signature for the diagnosis of kidney tumor types (Figure 3). The genes in this set may also be important for a differential antioncogenic therapy.
During the past years, the core unit at the DKFZ has generated a large amount of data on differentially expressed genes in human diseases. The challenge for the future is to use these data for the translation into the clinical setting. To this end, the gene expression signatures are screened for novel diagnostic and prognostic applications. At the same time, in collaboration with partners (e. g. SMP-Cell), our gene expression data give rise to high throughput cellular functional analyses and proteomics assays in order to understand disease pathomechanisms.
Furthermore, the technology is used for the identification of novel gene expression networks (see “Systematic Analysis of Functional Networks and Key Mediators”). Currently, biomedical research is at the stage of using microarrays as a routine tool and from this will give a considerable impact on the generation of hypotheses concerning disease pathomechanisms.


