
Introduction
Substantial bioinformatics and database expertise that works close together with the experimental projects is key to their mutual success, and thus a prerequisite for the progress of SMP-Cell. The ORF cloning pipeline of the cDNA Consortium forms the basis for subsequent functional genomics experiments in SMP-Cell and beyond. All steps of the ORF cloning process need to be managed with help of a dedicated information system in order to achieve and maintain high and consistent quality standards. In this line the cloned material additionally requires annotation after sequence validation and all information from ORF cloning and the annotation of cDNA sequences must be stored in an efficient data structure. The information structure on the one hand has to support the cloning process and on the other hand there it provides a direct link to data analysis and integration.
We develop and maintain laboratory information management systems (LIMS) to suit the different requirements and specifications of experimental processes and infrastructures. The data and information structure was built around the technical and experimental base. An in silico workflow was defined to transfer data in parallel with the progress of material. This data is integrated in a central database and complemented by relevant external data. This information from molecular functional gene analysis is made publicly available using the web-interface of the LIFEdb database (1) at http://www.lifedb.de.
 | |
| Fig 1: Consistent nomenclature of the products in ORF cloning. This allows to track any Gateway expression clone back to the respective entry clone, to the annotated ORF and to the template clone (or RNA). | |
Project Status
Standardized information flow
The ORF cloning process is distributed among five partners of SMP-Cell. This required a defined dataflow between these partners and the DKFZ, where all clones are maintained. Data about gene modeling, cDNA and ORF annotation are stored and edited in a MS Access database using an MS Access interface. To observe the status of the respective ORFs at all levels of processing we needed a standardized nomenclature (Fig. 1), which is also prerequisite for the tracing of data from subsequent application of clones in cellular functional gene analysis.
This nomenclature has proven to be essential for the unambiguous tracing of resources and information in SMP-Cell. Any data from cellular functional gene analysis can be related to the original clones and sequences it was based upon. The exchange and cross-platform comparison of information that has been obtained by collaboration partners is thus facilitated.
The cloning procedure begins with digital expressions that are related to physical material (i.e. mostly clones or RNA). We generate XML documents that contain all data for sets of ORFs and clones from the source in the central database. These documents are created by a client-server application which enables a user friendly selection of ORFs for processing by the different partners. After cloning is completed results-tables are sent back by the partners to the central database, again using XML documents.
 |
|
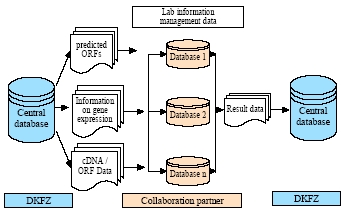
Fig 2:
The central database (blue) generates application XML-Files (white docs). These files contain relevant information to start the cloning procedure.
The cloning process (red) is carried out by several partners with help of dedicated databases. The results are fed back into the central database, again via XML-Flies (white doc).
Info-structured ORF cloning process
The processes leading from a gene model to a validated ORF resource are complex and manifold. We have selected the Gateway system to systematically clone ORFs in entry clones. These are sequence validated, and the ORFs are subsequently subcloned into a range of expression clones (1). This cloning schema has been adopted by all partners within SMP-Cell. The DKFZ delivers gene models and, when available, matching templates to the partners. There the ORF cloning takes place, and entry clones and accompanying information are returned (Fig. 2). All experimental processes are mapped to an electronic system in order to supervise these processes, to store relevant data and information, and to aid in the analysis and interpretation of results (Fig. 3). Bench-workers are thus enabled to trace every ORF and any construct that has been produced, and to decide on the next applicable steps that need to be performed to obtain a final product (i.e. entry clone).
 |
|
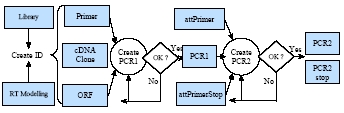
Fig 3a:
Workflow from gene modeling to the final PCR product that is ready for cloning.
 |
|
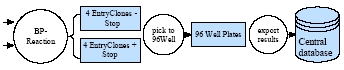
Fig 3b:
Workflow from the cloning of PCR products to the plate format in which the entry clones are stored and sequence validated. XML interfaces have been established for data transfer from local databases to the central database server at the DKFZ.
The development of a client/server application is in progress that allows to follow the workflow and to observe the progress of the OFR cloning. This application is tailored for remote use by the different partners for the distributed cloning processes. Next to the definition of a common nomenclature (Fig. 1), an XML standard format needed to be established that permits to import data of libraries, clones, and gene models, and to export any results from finished 96-well plates.
The software application SCISSORS serves two purposes and it consequently has two interfaces. Firstly, it serves as tracking database and administration tool for material and products. Secondly, this application is able to export data in XML files to transfer information from the central database to the collaboration partners and back.
Data integration and information structure All generated data and information from publicly available sources (2-5) is stored in separate databases on a central database server. The information is queryable across the different databases and comprises:
• annotation of genes and splice variants, ORFs, and cDNAs.
• data from ORF cloning and validation and the administration.
• experimental results from cellular functional gene analysis, i.e. protein localization and cellular assays.
• results from automated bioinformatic protein analyses (see PCE-S19T08)
• data from NCBI (Gene, UniGene), EBI (IPI, GO) and SIB (Swiss-Prot)
The data from the external sources is loaded by specialized applications and updated when new datasets become available. Some applications were developed and are in use e.g. for functional profiling (6, 7) or the software SCISSORS for the ORF cloning. We created a web interface for the uploading of external data (e.g. microscopic images from protein localization) which provides direct access to the central database server. Any data and information that is uploaded through this web interface becomes immediately accessible via the LIFEdb web-database (1).
Outlook
The currently established databases and applications offer a rapidly growing wealth of information from molecular functional gene analysis about genes, splice variants, ORFs, and cDNAs, and data about experimental results from cellular functional gene analysis. The design and implementation of software applications needs to be flexible in order to meet the increasing complexity of experiments and the organization of the information structure. The cloning of ORFs has become a high throughput operation and necessitates the transfer of information and resources between the different partners. The key application for the analysis will receive more features, “webservices” will be implemented with either Microsoft information server or with the Apache web server to substitute the current XML docs for SCISSORS.
Lit.: 1. Bannasch D et al. LIFEdb: A database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system. Nucleic Acids Res. 2004 32(1):D505-8. 2. Boeckmann B et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucl Acids Res. 2003 Jan 1;31(1):D365-70. 3. Maglott D et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005 Jan 1;33(1):D54-8. 4. Maglott D et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005 Jan 1;33(1):D54-8. 5. Rodriguez-Tome P. EBI databases and services. Mol Biotechnol. 2001 Jul;18(3):199-212. 6. Wiemann S et al. From ORFeome to biology: a functional genomics pipeline. Genome Res. 2004 14(10b):2136-44. 7. Arlt DH et al. Functional profiling: from microarrays via cell-based assays to novel tumor relevant modulators of the cell cycle. Cancer Res. 2005 65(17):in press.


