Introduction
Glioblastomas are the most common and most malignant primary brain tumors in human adult, accounting for about 50 % of all gliomas and 12-15 % of all intracranial tumors. Despite considerable progress in multimodality treatment, including neurosurgery, local radiation therapy, and chemotherapy, the prognosis of patients with glioblastoma is still poor, with a median survival time less than 1 year. Molecular genetic studies have identified a variety of chromosomal and genetic alterations in glioblastomas, with loss of chromosome 10 being present in up to 90 % of the cases. Chromosome 10 losses are far less common in lower grade astrocytic gliomas as compared to glioblastomas, suggesting that chromosome 10 carries glioblastoma-associated tumor suppressor genes (1). Previous studies have implicated the PTEN gene at 10q23 as an important tumor suppressor gene that is aberrant in affected in 25-40 % of primary glioblastomas (2). However, the absence of PTEN mutations in more than half of de novo glioblastomas and more than 90 % of secondary glioblastomas, i.e. glioblastomas that developed from a pre-existing lower grade glioma, clearly points to the presence of additional tumor suppressor genes on chromosome 10. By using loss of heterozygosity analyses, we and others have provided evidence for at least one additional glioblastoma-related tumor suppressor gene mapping to 10q24-qter, distal to PTEN (1, 2). Although several genes from this region, including DBMT1 (3), LGI1 (4), WDR11 (5), and MXI1 (6), have been proposed as glioblastoma suppressor gene, none of these candidates has been confirmed as being mutated in a significant fraction of tumors. Thus, the relevant glioblastoma suppressor gene(s) in 10q24-qter is/are still unknown.
Results/Project Status
In the first 3 months of our project, we have collected a core series of 100 primary gliomas with corresponding clinical and pathologic data. High molecular weight DNA and RNA have been extracted from these cases and are available for molecular analyses by the brain tumor network. At present, genomic and expression profiling using matrix CGH and oligonucleotide-based expression arrays is ongoing. The respective microarrays that are used for these experiments have been tailored to the specific questions concerning identification of glioblastoma-suppressor candidate genes on 10q24-qter.
With respect to the proposed project, we have already performed real time-RT PCR expression analysis of 15 genes located on 10q24-qter in a series of 34 de novo glioblastomas, 12 secondary glioblastomas, 11 anaplastic astrocytomas, 11 diffuse astrocytomas, and 5 glioma cell lines to identify novel candidate tumor suppressor genes that are downregulated in glioblastomas. The genes were either selected from the NCBI Human Genome Browser (www.ncbi.nlm.nih.gov) of from microarray expression profiling data of glioma cell lines treated with the demethylating agent 5-aza-2´-deoxycytidine and the histone deacetylase inhibitor trichostatin A. So far, we identified four (ADD3, EMX2, OAT, and INPP5A) genes that showed markedly reduced mRNA levels in substantial fractions of glioblastomas compared to diffuse astrocytomas and non-neoplastic brain tissue (Fig.1). Furthermore, treatment of glioma cell lines with 5-aza-2´-deoxycytidine and TSA
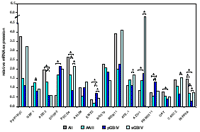
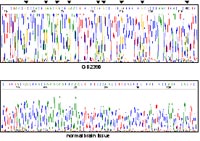
resulted in increase expression of these genes, suggesting that their transcriptional downregulation may be due to promoter hypermethylation. Subsequently, we investigated the promoter regions of these four genes by sequencing of sodium-bisulfite modified DNA and identified methylated CpG sites in the EMX2-promoter sequence (Fig. 2) in a fraction of glioblastomas (7/20, 35 %) with reduced expression of the gene.
In contrast, promoter hypermethylation is not the cause of the reduced expression of ADD3, OAT, and INPP5A, although treatment of glioma cell lines with demethylating agent restored expression of these three genes. EMX2 and ADD3 were also investigated for mutation and homozygous deletion by SSCP/heteroduplex analysis and genomic duplex PCR, respectively, but both genes exhibited neither genetic nor chromosomal alterations.
The molecular mechanisms leading to the the decreased expression of ADD3, INPP5A, and OAT as well as the functional significance of these genes, including EMX2, in glioma pathogenesis have to be clarified.
Outlook
The project aims at the identification as well as molecular and functional characterization of one or more novel glioblastoma suppressor genes from 10q24-qter. This aim will be addressed by using different approaches, including the development and application of a region-specific genomic BAC-array for matrix CGH analysis, as well as the development and application of a region-specific cDNA array for mRNA-based expression profiling. Candidate tumor suppressor genes identified by these approaches will be further characterized for mutations and/or epigenetic modifications in gliomas and subjected to functional analyses in vitro.
Lit.: 1. Reifenberger G, Collins VP. Pathology and genetics of astrocytic gliomas. J Mol Med. 2004 Oct;82(10):656-70. Review. 2. Knobbe CB et al. Pten signalling in gliomas. Neuro-oncol 2002, 4; 196-211. 3. Mollenhauer J. et al. DBMT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumors. Nature Genet 1997, 17; 32-39. 4. Chernova OB et al. A novel gene, LGI1, from 10q24 is rearranged and downregulated in malignant brain tumors. Oncogene 1998, 17; 2873-2881. 5. Chernova OB et al.,A novel member of the WD-repeat gene family, WDR11, maps to the 10q26 region and is disrupted by chromosome translocation in human glioblastoma cells. Oncogene 2001, 20; 5378-5392. 6. Wechsler DS et al. MXI1, a putative tumor suppressor gene, suppresses growth of human glioblastoma cells. Cancer Res 1997, 57; 4905-4912.


