Introduction
In NGFN-1 the Munich Center of the network Environmental Health has established a shared database to bring together the information of probands from different projects of the network. Information about DNA and phenotypes has been made available to every partner of the network. This project is continued in NGFN-2.
Results/Project Status
At the moment the database holds data of 48,000 patients and controls. The patients have been recruited by projects in Munich, Kiel and Berlin. Information and DNA has been made available to partners of the network.
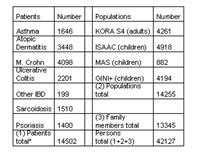
We have established a common database for the whole net Environmental Health. The database contains a minimum dataset on each patient/proband for overview purposes. The data presented to the users are available via the internet but only in highly aggregated form. As shown in table 1, this database meanwhile includes about 14 500 patients, 14 000 children and adults from the general population, and nearly 13 000 family members. Altogether data entries of more than 42 000 persons are available for case control studies and family-based studies. In addition to the use of this resource for the individual projects, the network has agreed to share the DNA of these subjects for common research activities of the partners in NGFN. It should be noted, that for each disease the available numbers of patients within this database are at least twice as many as the numbers from the individual projects.
Building on this implementation and database design, we have been asked to extend this concept on all clinical networks in NGFN-2, together with the SMP GEM. Partners hand in a preformatted set of data to the collection center. Real-time descriptive statistics are available online. Partners who contribute may in turn access other partners’ data and DNA. A proposal would be sent to the owners of the data and to the coordinating committee. All resulting genotypes will be fed into a joint DNA-database and are available for the whole network Environmental Health. The database in its current form is realised in MySQL with PHP.
Following the concepts of this existing database a NGFN-wide dataset that contains information about the existence of biosamples and clinical data is currently under development. The database has the objective to make the available sets of patients and probands transparent for all partners within the NGFN-2. Furthermore this database helps to improve reporting of the different NGFN units. To provide a better overview, all information is stored and presented only in an aggregated form; details of clinical or epidemiological information are not provided. Original data and biological samples always remain in the partners’ hands. The ownership of the data remains at the net projects.
To initiate and plan the realization of the database a workshop took place at the GSF in April 2005. Its result was the definition of a minimal set of variables which contains the required information about the existence of biosamples and clinical data. Next concepts the technical realization of the database have been developed. Meanwhile the concept is reviewed and the implementation has been started.
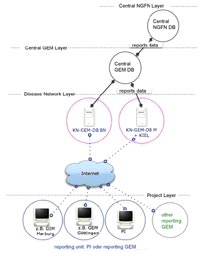
Reporting to different layers of system is planned according to the following schema (see also Figure 1):
1) Any funded NGFN-project in a disease network (KN) has to report its available data. The PI of the project together with the supporting GEM defines a contact person which is responsible for the regular reporting to the disease-net-GEM-databases (KN-GEM-DB) in Bonn (BN), Munich (M), and Kiel (KI).
2) The disease-net-GEM-databases (KN-GEM-DB) in BN, M, KI receive the data of the assigned projects and control the data for plausibility and completeness. The KN-GEM reports to the central GEM-database.
3) The central GEM-database receives the reports of the KN-GEM-DBs and controls for plausibility and completeness, as well as for duplicated entries. In the central GEM-DB the aggregation of the data is performed. The results are sent to the central NGFN-DB.
4) The central NGFN-DB is located at the DLR and serves as basis for the NGFN reporting. The reporting of the central GEM-DB is therefore the base for the external communication of the available data and new recruitments in the disease networks.
Outlook
The implementation of the system is ongoing. In short a first version will be made available and we will install the disease-net-GEM-databases in Munich and Bonn. After feeding in the data the system will provide overview information of the available patient data in the whole NGFN. This will facilitate the collaboration between the projects. It allows for multiple uses of available resources and identification of replication samples for new findings.
Lit.: 1. Weidinger, S., Klopp, N., Wagenpfeil, S, Rümmler, L., Schedel, M., Kabesch, M., Schäfer, T., Darsow, U., Jakob, T., Behrendt, H., Wichmann, H.E., Ring, J., Illig, T.: Association of a STAT 6 haplotype with elevated serum IgE levels in a population based cohort of white adults. J Med Genet 41(9), 658-63 (2004); 2. Weidinger, S., Klopp, N., Rummler, L., Wagenpfeil, S., Novak, N., Baurecht, H.J., Groer, W., Darsow, U., Heinrich, J., Gauger, A., Schäfer, T., Jakob, T., Behrendt, H., Wichmann, H.E., Ring, J., Illig, T.: Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J Allergy Clin Immunology 116, 177-84 (2005); 3. Weidinger, S., Klopp, N., Rümmler, L., Wagenpfeil, S., Baurecht, H.J., Gauger, A., Darsow, U., Jakob, T., Novak, N., Schäfer, T., Heinrich, J., Behrendt, H., Wichmann, H.E., Ring, J., Illig, T. for the KORA study group: Association of CARD15 Polymorphisms with Atopy-related Traits in a Population-Based Cohort of Caucasian Adults. Clin Exp Allergy 35, 866-872 (2005); 4. Weidinger S., Klopp N., Wagenpfeil S., Rümmler L., Baurecht H.J., Fischer G., Holle R., Jakob T., Darsow U., Schäfer T., Behrendt H., Ring J., Illig T.: Association study of mast cell chymase polymorphisms with atopy. Allergy in press (2005). 5. Schedel, M., Carr, D., Klopp, N., Woitsch, B., Illig, Th., Stachel, D., Schmid, I., Fritzsch, C., Weiland, S.K., von Mutius, E., Kabesch, M.: A signal transducer and activator of transcription 6 haplotype influences the regulation of serum IgE levels. J. Allergy Clin. Immun. 114, 1100-1105 (2004); 6. Vollmert, C., Illig, T., Altmüller, J., Klugbauer, S., Loesgen, S., Dumitrescu, L., Wjst, M.: Single nucleotide polymorphism screening and association analysis - exclusion of integrin ß7 and vitamin D receptor (chromosome 12q) as candidate genes for asthma. Clin. Exp. Allerg. 34, 1841-1850 (2004); 7. Hysi, P., Kabesch, M., Moffatt, M.F., Schedel, M., Carr, D., Zhang, Y., Boardman, B., v. Mutius, E., Weiland, S.K., Leupold, W., Fritzsch, C., Klopp, N., Musk, A.W., James, A., Nunez, G., Inohora, N., Cookson, W.O.: NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet 14(7), 935-41 (2005); 8. Kormann, M.S.D., Carr, D., Klopp, N., Illig, T., Leupold, W., Fritzsch, C., Weiland, S.K., v. Mutius, E., Kabesch, M.: G-Protein-coupled Receptor Polymorphisms Are Associated with Asthma in a Large German Population. Am J Respir Crit Care Med 171, 1358-1362 (2005)


