
Introduction
Breast cancer was estimated to be the most frequent malignancy among women in the western world in 2004. Although much effort has been put into designing new and sophisticated drugs, classical chemotherapeutic agents such as Doxorubicin (DXR), Cyclophosphamide (CPA) or Taxotere™ (TXT) are still widely employed in the clinic. A major problem of most of the current therapies is the intrinsic or acquired drug resistance of cancer cells, which leads to relapse and metastatic progression. Therefore a comprehensive characterisation of genes contributing to a drug resistance phenotype is needed. Genetic comparison of apoptosis-resistant with apoptosis-sensitive breast cancer cells will lead to the identification of genes responsible for resistance to apoptosis.
In order to identify genes which are involved in the signal transduction of resistance mechanisms we treated breast cancer cell lines with different chemotherapeutic drugs. The gene expression pattern of surviving clones was analysed with DNA-Macroarrays and compared to that of the original cell line.
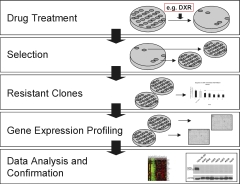 | |
| Fig 1: Experimental procedure | |
The contribution of each single gene to apoptosis resistance in general and to a certain resistance mechanism can be analysed in vitro as well as in vivo. Functional inhibition of these gene products will enable us to develop new strategies towards solving the apoptotic blockade and to drive cells actively into programmed cell death.
Results
Gene expression profiling of apoptosis-sensitive and -resistant breast cancer cell lines
Chemotherapeutics (Doxorubicin, Cyclophosphamide, etc.) can trigger apoptosis in breast cancer cell lines. Addition of these cytotoxic agents in appropriate doses to certain breast cancer cell lines results in the survival of a few cells. These surviving clones can be selected for apoptosis resistance by increasing concentrations of the chemotherapeutic agent. We generated about 50 clones treated with either Doxorubicin, Cyclophosphamide or Taxotere™. We determined the apoptosis rate after inducing cell death with the corresponding chemotherapeutic agent in these clones. More than 65% of the clones of different breast cancer cell lines showed significant decrease in apoptosis.
The resistant clones were subjected to gene expression analysis by Macroarrays. Because of the well defined gene expression phenotype of the original breast cancer cell line, changes in gene expression in resistant clones can be monitored very precisely. The cDNA Array Screeningsystem of the MPI-MB department has been established a couple of years ago and has been further developed since. This macroarray system comprises rationally selected gene families and signalling pathway components. It allows the analysis of all known protein kinases and phosphatases as well as numerous marker genes of apoptosis and tumorigenesis.
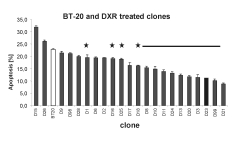
Fig 2: Apoptosis resistant clones BT-20 breast cancer cells selected with DXR. Cells were PI stained and analyzed with a FACS. Cells in the subG0/1-phase were regarded as apoptotic. (p<0,05)
Until now 150 Arrays of different breast cancer cell lines and their chemotherapeutic resistant clones have been analysed. Genes with altered gene expression in apoptosis resistant or sensitive cells can thereby be identified. Analysis of the expression level of a selected subset of genes of interest revealed over 100 genes that are at least 2 fold up- or downregulated in the clones as compared to the original cell line. Among these genes are well known anti- and proapoptotic enzymes as well as enzymes that have not been linked to apoptosis before. The involvement in chemoresistance of about 20 genes that have been upregulated in the resistant clones will be further analysed. These proteins act in different antiapoptotic pathways and promote survival of the cell when overexpressed. Several genes were repressed in apoptosis resistant clones including genes with known proapoptotic function and others whose functions in apoptosis need to be elucidated.
Comparison of genes with increased expression in clones of the same breast cancer cell line BT20 but treated either with DXR, CPA or TXT revealt a couple of differentially expressed. These genes are now further characterised to analyse their specific contribution to the chemoresistance phenotype.
Outlook
Analysis of identified markers of apoptosis resistance - new targeted approaches for therapiesThe vast amount of biological material of the parental cell line as well as of the resistant clones allows us to analyse the specific contribution of single genes to the resistance phenotype and thereby validate them as targets for further drug development. Proteins with increased gene expression could be inhibited by small molecule drug inhibitors or their gene expression could be knocked down in siRNA-experiments. To restore decreased gene expression, infection of resistant clones with relevant constructs could be performed. In addition, these cell lines would be suitable for the functional analysis of proteins via mutant forms to determine their influence on apoptosis. With the aid of these and other methods it will be possible to discover new targets for the interference with apoptosis resistance.
Diagnosis of chemoresistance in tumors
Because of the evolutionary development of cancer cells, every tumor has its own characteristic gene expression profile that is defining the hallmarks of cancer. The goal of this method is to transfer the information obtained from the cellgenetic approach described above to primary tumors. In this way we intend to establish a system for the diagnosis of apoptosis resistance on the basis of molecular determinants (apoptosis pathway signature).
Furthermore, this model could serve as groundwork for the development of antiapoptotic therapies.
Our functional genomics strategy opens up the opportunity to tailor therapies to the individual tumor or patient. A combination of classical chemotherapy and targeted drugs could then be used to prevent the development of chemoresistance.
In this project a comprehensive characterisation of genes contributing to a drug resistance phenotype is intended. Genetic comparison of apoptosis-resistant with apoptosis-sensitive breast cancer cells is planned to lead to the identification of genes involved in the formation of resistance to apoptosis in breast cancer. The functional inhibition of corresponding gene products will enable us to develop new strategies towards breaking the apoptotic blockade and to drive cells actively into programmed cell death.


