
Introduction
About 80% of all cases of colorectal cancer (CRC) show mutations in the tumor suppressor gene adenomatous polyposis coli (APC). These mutations occur early in the development of CRC and are linked to the hyperproliferation of normal colonic epithelium leading to the formation of adenoma. APC has been suggested to act as a gatekeeper whose loss of function allows the consecutive acquisiton of further mutations in oncogenes and tumor suppressor genes such as ras or p53 and thus promote the further progression of the disease (Kinzler, Vogelstein, Cell. 87(2),159-70, 1996). Functionally, mutations of APC result in the stabilization of the cytoplasmic protein b-catenin which accumulates in the nucleus and combines with transcription factors of the TCF/LEF-1 family (below referred to as TCF). In some of the CRC that have wild-type APC, and in a variety of other cancer types, dominant mutations of b-catenin are present, qualifying b-catenin as an important oncogene. Under physiological conditions b-catenin is stabilized by Wnt signaling, and TCF/b-catenin complexes lead to activation of Wnt target genes. Several of these Wnt targets are also implicated in tumorigenesis, such as cyclinD1 and c-myc, and various matrix metalloproteinases. It is likely that more Wnt pathway-regulated genes that mediate the full blown cancer phenotype have yet to be discovered. Transcriptional alterations of these genes should be apparent in comparisons of transcriptomes of CRC and normal mucosa.
Our group has a longstanding record in the molecular analysis of b-catenin signaling (1). We have identified key components in this pathway and contributed to the concept of b-catenin-induced tumor formation. In particular we have discovered the interaction of b-catenin with LEF-1, a member of the TCF family (2), and with conductin, a cytoplasmic protein that also binds to APC and is essential for degradation of b-catenin (3, 4).
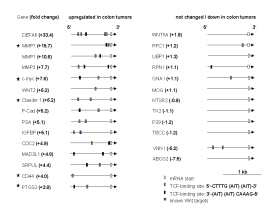 | |
| Fig 1: Presence of TCF consensus binding sites in 1 kb of upstream region of genes upregulated in CRC (left) or genes unchanged/downregulated (right) as compared to normal mucosa. Note the prevalence of TCF sites in genes that are upregulated in CRC indicating putative Wnt targets. Known Wnt target genes are indicated by asterisks. Genes were randomly selected from the Affymetrix analysis list provided by F. Rödel, Erlangen. | |
Project Status
Microarray analysis performed at our research site has identified a number of genes genes that are differentially regulated between colorectal tumors and normal colonic mucosa. Among the genes upregulated in CRC (202 genes, ³ 3 fold) are known targets of the Wnt pathway but also a large number of other genes that have not yet been linked to Wnt signaling. Our bioinformatic analysis shows that TCF/b-catenin binding sites indicative of Wnt target genes are significantly overrepresented in genes upregulated in CRC as compared to genes which are not changed or downregulated (2.1 vs 0.4 binding sites per 1kb of upstream promoter region, Fig. 1).
By microarray analysis we could identify 208 genes to be downregulated (³ 3 fold) after expression of a dominant-negative TCF-4 mutant in a colorectal cancer cell line (DLD1). By matching genes upregulated in CRC with those downregulated by dominant-negative TCF we could narrow down the list of potential Wnt targets to 151 genes. Thus the spectrum of genes regulated by Wnt signaling in CRC appears to be much broader than suggested by the current list of known Wnt target genes. Since activation of the Wnt pathway is an essential and early step of colorectal tumorigenesis these novel target genes are excellent candidates for diagnosis and drug targeting.
In the analyses we noticed that several components of the Wnt pathway are upregulated in the tumors. For instance, we could show that conductin, a negative regulator of the pathway, is upregulated in CRC and liver tumors and is a target gene of TCF/b-catenin (5). We have also analysed the nuclear factors pontin and reptin, which were shown to interact with b-catenin and to differentially regulate its activity: while pontin activates transcription by TCF/b-catenin, reptin represses transcription. We have cloned the promoters of both genes and found that pontin, but not reptin promoter activity is directly increased by TCF/b-catenin. This indicates that a positive feedback loop exists in CRC which superactivates the Wnt pathway through the selective upregulation of pontin. Of note, pontin is a cofactor of the c-myc protein and might thus also act further downstream in the Wnt signaling cascade (Kurniati et al., Manuscript in preparation).
We also found that several genes with a function in mitosis (e.g. cdc2, mad2) are upregulated in CRC. Specifically we have analysed the mad2 gene and could show that its promoter is activated by TCF/b-catenin. Since mad2 is a component of the mitotic spindle check point machinery which controls the fidelity of chromosomal seggregation our data indicate that Wnt signaling directly impinges on mitotic processes. This finding is of importance given the frequent observation of chromosomal instability in CRC.
Rationale
We assume that many of the genes transcriptionally altered in CRC as compared to normal mucosa are not causally involved in cancer formation but are rather indirectly regulated as a general consequence of tumor growth. To distinguish the “important” genes from the “bystanders” we concentrate on genes directly regulated by aberrant activation of the Wnt pathway.
Identification of candidate Wnt target genes
As exemplified above, our strategy is to compare changes in gene expression in CRC vs. normal mucosa to those in cellular systems where we can inhibit or activate Wnt signaling in a controlled manner. Genes that match in such an analysis will be further studied in promoter and functional experiments. We apply the following criteria for narrowing down genes from our initial comparison (i.e. the 151 genes upregulated in CRC and downregulated by dom.-neg. TCF):
(i) Likelihood to be involved in a signaling pathway or major regulatory step in cancer from known function or sequence.
We focus on genes with a putative role in signal transduction events, such as growth factors, kinases, phosphatases, transcription factors or genes with a putative function in other tumor-related phenotypes, e.g. invasion, angiogenesis, and cell cycle regulation. Additional interesting candidates are components of the Wnt pathway, which might have negative or positive feedback roles as seen with conductin or pontin. There might also be genes that are upregulated in CRC but - due to cell line specificities - not repressed by dom.-neg. TCF in the DLD1 system. To avoid neglecting such genes we will also perform microarray analysis of alternative cellular models for Wnt pathway regulation. For instance we have established previously that Wnt conditioned medium can induce stabilization of b-catenin and expression of conductin in breast carcinoma cells (5).
(ii) Regulation of promoter activity by TCF/b-catenin complexes.
For analysis of direct transcriptional regulation of a given gene by TCF/b-catenin we have cloned promoter sequences by genomic PCR and generate luciferase reporter constructs. The putative transcription start sites will be taken from the literature, if available, or determined by computer-assisted alignment of EST clones with genomic sequences. The reporters are tested for activation by TCF/b-catenin or for repression by dominant-negative TCF. Promoters which show clear regulation are mutated at the putative TCF sites to validate that the regulation depends on direct interaction with TCF. In addition chromatin IP assays will be performed to demonstrate TCF/b-catenin binding to promoters in vivo. By these analyses we expect to to obtain criteria for further selection of genes for functional assays.
We will also analyze genes that are downregulated in CRC and upregulated by dom.-neg. TCF. These genes are thought to be indirectly controlled by Wnt signaling but might nevertheless have important functions. A paradigm for this type of regulation is the p21CIP gene which is repressed by c-myc in response to Wnt signaling.
Outlook
Functional analysis
For the functional analysis we will either upregulate the encoded proteins by exogenous expression of the respective cDNAs or suppress the expression of endogenous proteins by RNA interference technology. As read-out systems we will use assays that allow to determine effects on cell proliferation and apoptosis on a single cell basis (e.g. BrdU incorporation and staining with annexinV, repectively).
We will also determine whether individual genes can affect invasiveness. Here we will use a collagen invasion assay where transfected cells can be traced after co-transfection of a GFP marker plasmid. For selected genes, stable transfectants will be generated that will also allow in vivo analysis of tumor growth and metastasis formation in collaboration with the CancerNet project “Animal tumor models for the evaluation of candidate gene function in tumor metastasis” (J. Sleeman).
In specific cases it might also be possible to interfere with protein function with blocking antibodies or to test the activity of secreted proteins by use of recombinant proteins or conditioned medium of transfected cells.
A specific focus will be on the role and regulation of genes involved in mitosis or regulation of S-Phase which we have frequently found in our microarray analyses. Here we have established a variety of assays such as mitotic checkpoint analysis, FISH for analyzing chromosomal content, and assays for microtubule dynamics. Evidently, in the end we will have to focus on a rather limited number of attractive candidates for detailed analysis.
Expression analysis in CRC
The protein expression pattern of individual candidates will be determined for retrospective analysis of paraffin-embedded tumor material, and with the project Brabletz/Kirchner for specific analysis at the invasive front of primary tumors and in metastases.
Lit. :1. Huelsken J, Behrens J. The Wnt signaling pathway. J Cell Sci. 2002 Nov 1;115(Pt 21):3977-8. 2. Behrens J et al, v. Kries J.P., Kühl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. (1996) Functional interaction of b-catenin with the transcription factor LEF-1. Nature 382: 638-642. 3. Behrens J et al (1998) Functional interaction of an axin homolog, conductin, with b-catenin, APC, and GSK3b. Science 280: 596-599. 4. Schwarz-Romond T et al (2002) The ankyrin repeat protein Diversin recruits casein kinase Ie to the b-catenin degradation complex and acts in both canonical Wnt and and Wnt/JNK signaling. Genes Dev. 16:2073-84. 5. Lustig B et al (2002) Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell Biol. 22: 1184-93.


