Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignancy in childhood. Major advances have been made in the treatment of childhood ALL in the last several decades, based on the identification of prognostic markers and the development of risk-adapted treatment strategies. However, about 25% of patients still fail therapy and surviving patients often suffer from significant toxicities. Therefore, an improvement of the assessment of an individual’s risk of relapse is necessary in order to adapt treatment and to reduce toxicity and improve treatment outcome.
In our current clinical trial ALL-BFM 2000, risk-adapted treatment stratification (standard- (SR), intermediate- (IR), or high-risk (HR)) is performed using cytogenetic markers (t(9;22), t(4;11)) or their molecular equivalents (BCR-ABL and MLL-AF4), and the in vivo response to treatment. Response is assessed cytomorphologically by the initial cytoreduction (blast reduction in peripheral blood after 7 days of treatment with prednisone and one intrathecal application of methotrexate: “prednisone response”; blast clearance from bone marrow after induction therapy on treatment-day 33), or molecularly by measurement of MRD on treatment-day 33 and after induction consolidation at week 12. Measurement of MRD is based on the detection of clone-specific immunoglobulin and T-cell receptor gene rearrangements by PCR amplification and generally reaches sensitivities of 10-4 - 10-5.
The detection of one of the cytogenetic markers (t(9;22), t(4;11)), a poor prednisone response and a poor blast clearance from bone marrow on treatment-day 33 as well as a high MRD load after induction consolidation qualify the patients for the HR-group. HR-patients are treated using a more intensive chemotherapy, including myeloablative treatment with allogeneic hematopoietic stem cell transplantation (SCT). However, this group still comprises the highest rate of relapses. The severe side effects of treatment not only lead to severe acute toxicity and life-long sequelae but also to a significant rate of treatment related mortality. To avoid the dilemma of overtreatment (e.g. by myeloablative chemotherapy and SCT) in some patients as much as the undertreatment of all those patients who ultimately will relapse, further improvement of risk assessment procedures for treatment stratification especially within this HR-group is necessary.
It has recently been shown that gene expression profiling using microarrays can accurately identify genetic subtypes of pediatric ALL. This proves that, in principle, expression signatures can be identified for this entity. However, in the vast majority of cases these signatures are not yet sufficient to serve as reliable prognostic markers, e.g. to predict the risk of relapse for an individual patient.(4-11) Therefore, additional analyses are necessary that better define specific and prognosis associated gene expression profiles.
Project Status
To determine whether gene expression profiling of initial leukemic samples can distinguish good from poor molecular treatment response as assessed by the measurement of MRD after induction therapy and induction consolidation, we compared the expression signatures of 10 ALL samples with a good to those of 10 samples with a poor molecular response. A supervised approach selected 148 genes that exclusively distinguished good from poor treatment response, some of which are associated with DNA repair, signaling, cell cycle control, apoptosis and oncogenesis. Results were confirmed by quantitative RT-PCR with correlation coefficients between 0.823 and 0.907. Prediction analysis of microarrays (PAM) using randomly selected samples as a training set and the remaining samples as a test set revealed an accuracy of 84%. In conclusion, we identified several biologically interesting genes, which might explain differences in treatment response in childhood ALL. As treatment response could be predicted with a high accuracy, our study encouraged gene expression profiling in a larger series of patients with the aim to develop new clinically relevant tools for a further improvement of treatment stratification in childhood ALL.
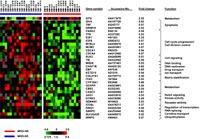
Based on our pilot data, we are now prospectively investigating in a large patient population of our ongoing trial whether gene expression profiling of initial leukemic samples by microarray technology can be used as an additional tool to further improve treatment stratification, especially within the HR-group of patients. We are also trying to find out whether there are common gene expression patterns of high risk ALL independent of the parameters used for stratification into the HR-group. An additional aim of our ongoing project is to further characterize genes which are correlated with treatment response and outcome, and to identify those which may be utilized for a specific targeted therapy.
Another important issue for our work is standardization of experimental proecedures. A fruitful cooperation exists with the cooperating groups within the NGFN performing gene expression analyses in childhood ALL, e.g. the group of C. Hagemeier, K. Seeger and W. D. Ludwig in Berlin and the group of A. Kulozik in Heidelberg. Together we developed standards in sample and RNA preparation, an indispensable pre-condition for the exchange of material. Furthermore, we are actively exchanging data to optimize our analyses.
Outlook
To define the molecular signatures of sensitivity or resistance in childhood ALL, we are currently validating our results in a prospective study with a large series of patients. In the future, gene expression profiling may eventually become a useful, clinically relevant tool for treatment stratification in the early course of childhood ALL and may contribute to an improvement in treatment outcome by defining the optimal therapy for each patient.
Lit.: 1. Schrappe M, Camitta B, Pui CH, Eden T, Gaynon P, Gustafsson G, et al. Long-term results of large prospective trials in childhood acute lymphoblastic leukemia. Leukemia 2000;14(12):2193-4. 2. Fine BM, Stanulla M, Schrappe M, Ho M, Viehmann S, Harbott J, Boxer LM. Gene expression patterns associated with recurrent chromosomal translocations in acute lymphoblastic leukemia. Blood 2004;103(3):1043-9. 3. Cario G, Stanulla M, Fine BM, Teuffel O, Neuhoff NV, Schrauder A, Flohr T, Schafer BW, Bartram CR, Welte K, Schlegelberger B, Schrappe M. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood 2005;105(2):821-6.


