Introduction
The analysis of chromosomal translocations in hematopoietic malignancies has yielded great insights into the pathogenesis of these diseases. Balanced chromosomal translocations can either lead to the formation of a fusion gene (e.g. the BCR/ABL1 resulting from the t(9;22) in chronic myelogenous leukemia) or to deregulation of gene expression through the juxtapposition of a strong promoter (e.g. upregulation of CMYC from the t(8;14) in Burkitt’s lymphoma). The genes that are found at the breakpoints of these translocations are often of crucial importance in normal hematopoietic differentiation and proliferation.
The focus of this project is the CALM/AF10 fusion gene, which results from the t(10;11)(p13;q14) translocation (1, 2). This translocation and the corresponding fusion gene are found in patients with acute myeloid leukemia (AML), T-cell acute lymphoblastic leukemia (T-ALL), and in malignant lymphomas. The AF10 gene is located on chromosome 10 band p13 and the CALM gene is found on chromosome 11 band q14. The CALM/AF10 fusion transcript encompasses nearly the complete open reading frames of both the CALM and AF10 genes, whereas the reciprocal AF10/CALM fusion essentially encodes for a truncated AF10 protein (fig. 1).
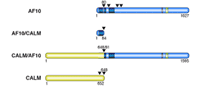
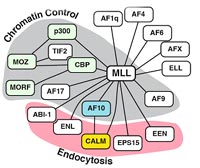
The AF10 gene was originally cloned as the fusion partner of the MLL gene, the human homologue of the Drosophila trithorax gene. The MLL/AF10 fusion is found mainly in AML. MLL has been described as the fusion partner of more than 40 other genes in various types of leukemia. Interestingly, an MLL/CALM fusion was described recently in a case of AML. The various fusions are depicted in figure 2.
AF10 is a putative transcription factor and found mainly in the nucleus of cells. In contrast, its fusion partner CALM (Clathrin Assembly Lymphoid Myeloid leukemia gene) is a protein, which is mainly found in the cytoplasm where it is associated with the Golgi apparatus and the cell membrane. AF10 is expressed at lower levels in the cells compared to CALM, which is ubiquitously expressed and as, a clathrin assembly protein, is involved in clathrin-mediated endocytosis.
CALM/AF10 is an interesting leukemic fusion for several reasons: a) it is found in several subtypes of leukemia and in lymphoma; b) quite often the t(10;11) is the sole cytogenetic abnormality in these leukemias suggesting an important role for CALM/AF10 in the development of the leukemia; c) it is one of the few fusion genes in which a fusion partner of MLL is independently fused to a third gene; d) retroviral transduction of murine bone marrow cells with a CALM/AF10 expressing retrovirus and subsequent transplantation of the CALM/AF10 expressing bone marrow cells into lethally irradiated mice causes an aggressive leukemia with short latency of only 9 to 15 weeks. These experiments demonstrated that CALM/AF10 is one of the strongest leukemogenic fusion genes that we know.
We thus set out to identify the target pathways that are affected by the CALM/AF10 fusion protein and which are presumably critically altered in leukemogenesis not only in the case of CALM/AF10 positive leukemias but most likely also in other leukemias.
Results
Two approaches were chosen to identify target pathways of CALM/AF10: a) the analysis of global gene expression levels in CALM/AF10-positive patients and in cell line models with an inducible CALM/AF10 gene to identify CALM/AF10 target genes; and b) identification of CALM and AF10 interacting proteins.
CALM/AF10 Target Genes
The CALM/AF10 fusion is not very common in AML, where it is found in less than 1% of the cases. The fusion is slightly more common in T-ALL and can be found in up to 20% of T-ALL cases with T-cell receptor ### rearrangement.
We analyzed samples from 13 patients with different types of leukemia and a t(10;11), in which an MLL rearrangement had been excluded: 5 cases of AML, 6 cases of T-ALL, 1 case of AUL and 1 case of acute biphenotypic leukemia. The samples were analyzed for the presence of the CALM/AF10 and AF10/CALM fusion transcipts by RT-PCR and sequence analysis. All these patients were found positive for the CALM/AF10 fusion. In addition, we analyzed a series of 29 patients with T-ALL with TCR #d rearrangement. Among these patients, four (4/29) were positive for CALM/AF10 transcripts, indicating a high incidence of CALM/AF10 fusions in this group of leukemia. We found three different breakpoints in CALM at nucleotide 1926, 2091 and at nt 2064 of CALM. In AF10 four breakpoints were identified: at nucleotide position 424, 589, 883, and 979. In seven patients it was also possible to amplify the reciprocal AF10/CALM fusion transcript. There was no correlation between disease phenotype and breakpoint location. Ten CALM/AF10 positive patient samples were subjected to expression profiling using oligonucleotide microarrays representing 33,000 different genes (U133 set, Affymetrix). The expression data obtained from the CALM/AF10 positive patients were compared to the expression profiles of several other groups of leukemia (e.g. t(8,21) positive AML samples). This analysis revealed high expression levels of the polycomb group gene BMI1, the homeobox gene MEIS1 and the HOXA cluster genes HOXA1, HOXA4, HOXA5, HOXA7, HOXA9, and HOXA10. The overexpression of HOX genes seen in these CALM/AF10 positive leukemias is reminiscent of the pattern seen in leukemias with rearrangements of the MLL gene (fig. 3), normal karyotypes or complex aberrant karyotypes. Overall there were many more genes repressed in CALM/AF10 positive leukemias than activated. The Drosophila AF10 homologue can act on polycomb group responsive elements. It is thus conceivable that the CALM/AF10 fusion protein acts in a dominant negative fashion on wild type AF10 function, relieving the repression that is presumably normally exerted by AF10 on the expression of HOX genes.

One drawback of analyzing the expression profile of leukemic patient samples is that we lack the expression profiles of the corresponding normal cells. Therefore it is very difficult to know whether expression differences seen in comparison to an arbitrary „normal“ control, eg. normal bone marrow, are due to the action of the transforming fusion gene or to the fact that different differentiation stages and lineages of hematopoietic cells are compared to each other. In addition, the action of the fusion protein will have probably caused many secondary, tertiary etc. changes in gene expression so that the expression signature of the direct target genes of the fusion gene will be greatly masked. In order to identify the direct target genes of the CALM/AF10 fusion gene, we have established a cell line in which the expression of the fusion gene can be induced. Using this system, genes that respond to the expression of CALM/AF10 within a few hours can be identified. Expression profiling of this system is currently in progress.
CALM and AF10 Interacting Proteins
As a second approach to identify pathways that are critically altered by the presence of the CALM/AF10 fusion protein we screened for CALM and AF10 protein interactors using the yeast two hybrid approach.
Several CALM interactors were found: TSG101 (tumor susceptibility gene 101), CATS (CALM interacting protein expressed in Thymus and Spleen), MCM2 (minichromosome maintenance 2), CALM, PCBP1 (poly(rC) binding protein 1), FHL2 (four and a half LIM domains 2), DPP7 (dipeptidyl-peptidase 7) and several other interactors.
The identification of a protein interaction in the yeast two hybrid screen is only a first step in studying protein interactions. The interactions have to be confirmed with other methods and their significance has to be analyzed by a variety of other techniques. Of the above named proteins and protein interactions one focus of our work is on the CATS protein.
The CATS gene had not been described before. Multiple tissue Northern blot analysis using a CATS probe showed a 1.6 kb transcript which was predominantly expressed in spleen and thymus and to a lesser extent in small and large intestines. Sequence analysis of a full length CATS cDNA revealed an open reading frame of 238 or 248 amino acids. The amino acid sequence of CATS shows no significant homologies to other proteins in the database. The CATS gene is located on chromosome 17, has at least 5 exons and spans approximately 6 kb.
GST-pulldown and co-immunoprecipitation experiments with over-expressed CATS protein and native CALM in HEK293T cells confirmed the CALM-CATS interaction found in the yeast system. Several monoclonal antibodies against the C-terminus of human CATS were raised (in cooperation with Dr. Kremmer, GSF). These antibodies recognize both the human and the murine CATS protein. Using these antibodies we could show a high expression of CATS in different human leukemia, lymphoma and solid tumor cell lines, as well as in normal proliferating cell lines (HEK293 and WI38), but not in normal non-proliferating T-cell lines (TYRF8 and JB4). Using protein lysates from cell cycle synchronized cells (Hela and U2OS) a clear cell cycle dependent regulation of CATS protein levels was demonstrated. Moreover, Western blot analysis showed that serum stimulation of serum starved T98G glioblastoma cells leads to increased expression of CATS.
Transient transfection studies revealed that CATS is localized mainly to the nucleus in nodular structures. Coexpression of CFP-CATS with YFP tagged nucleolar proteins (nucleostemin) showed that CATS is found predomi-nantly at the nucleoli. Coexpression of CFP-CATS with YFP-CALM or YFP-CALM/AF10 was able to markedly increase the nuclear localization of both CALM and the CALM/AF10 fusion protein. This effect of CATS is stronger on the YFP-CALM/AF10 fusion protein than on the CALM protein.
Our results indicate that the subcellular localization of CALM and CALM/AF10 could depend in part on the presence of CATS with a greater fraction of CALM or CALM/AF10 being present in the nucleus in cells with high CATS expression (e.g. lymphoid cells). High expression of CATS in proliferating cells and in tumor cells together with its nucleolar localization suggest that CATS is involved in controlling cell proliferation. The CALM-CATS interaction might thus play an important role in CALM/AF10 mediated leukemogenesis.
The AF10 interaction screen revealed interactions of AF10 with DNA repair associated proteins (DDB1: damage-specific DNA binding protein1; RAD23A: UV excision repair protein RAD23 homolog A) and with a zinc finger transcription factor which is critical for the development of all lymphoid lineages.
Outlook
Our approach has yielded valuable insight into possible pathways through which CALM/AF10 mediated leukemogenesis might operate (HOX gene deregulation, nucleolar function, disturbed subcellular localization of important proteins, DNA repair, disturbed lymphoid differentiation). It was recently shown that AF10 interacts with the histone methyltransferase hDOT1L (3). In collaboration with Dr. Xu, Shanghai, we could recently show that CALM/AF10 positive patient samples show global changes in histone methylation. These findings will guide our future research and should eventually lead to a better understanding of the pathogenesis of these aggressive CALM/AF10-positive leukemias and possibly of other leukemias as well.
Lit.: 1. Dreyling et al. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. PNAS 93:4804-4809 (1996). 2. Bohlander et al. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia 14:93-99 (2000). 3. Okada et al. hDOT1L links histone methylation to leukemogenesis. Cell 121:167-178 (2005).


