Introduction
Experimental data have demonstrated that in most cases single genetic alterations such as fusion genes (e.g. AML1-ETO) or mutations (e.g. FLT3-LM’s) are not able to induce leukemia in murine in vivo models, pointing to the relevance of collaborating genetic alterations for leukemogenic transformation. The length mutation of the tyrosine receptor kinase FLT3 is one of the most frequent genetic alterations in acute myeloid leukemia. However, constitutive expression of this genetic alteration in normal murine bone marrow cells by retroviral gene transfer failed to induce overt leukemia in transplantated mice on its own. The goal of this project was to determine collaborating events in FLT3-LM positive AML based on data sets of cytogenetically and molecularly well defined AML cases and cDNA microarray analyses.
Project Status
In a first step the project concentrated on potential cytogenetic genetic events collaborating with the FLT3-LM. For this we followed the model of pathogenesis of acute leukemia, in which two groups of genetic alterations, one affecting transcriptional regulation and hematopoietic differentiation, the other altering signal transduction cascades associated with cell proliferation, collaborate in inducing acute leukemia. One candidate for collaboration with FLT3-LM is the fusion gene AML1-ETO, which represents the class of alterations affecting transcriptional regulation and is recurrently found in patients with FLT3-LM positive AML: when 135 patients with AML1-ETO were screened for activating mutations in the receptor tyrosine kinases FLT3 and KIT as well as in NRAS (KITD816, NRAS codon 12/13/61), activating mutations were detected in 38 patients (28.1 %) and included mutations in the receptor tyrosine kinases FLT3 (11 patients) or KIT (25 patients in total) or in NRAS (13 patients). In contrast no MLL-PTD (partial tandem duplication) mutations were detected in 87 samples subjected to this analysis. These data demonstrated that FLT3 LM are a recurrent finding in patients with AML1-ETO positive AML.
To test the functional significance of the association of FLT3-LM with the AML1-ETO fusion gene we utilized the murine bone marrow (BM) transplantation model. MSCV-based retroviral constructs carrying the AML1-ETO cDNA upstream of an IRES-GFP cassette or the FLT3-LM cDNA upstream of an IRES-YFP cassette were generated to transduce and track hematopoietic cells expressing AML1-ETO (GFP+), FLT3-LM (YFP+) or both AML1-ETO and FLT3-LM (GFP+/YFP+) in vitro and in vivo. In order to investigate the impact of expression of FLT3-LM and AML1-ETO individually on primary primitive hematopoietic progenitor cells we performed the Colony-Forming Spleen Assay (CFU-S): bone marrow cells transduced with the AML1-ETO/GFP or FLT3-LM/YFP vector or both vectors were highly purified 96 hours after the start of infection by FACS. Their ability to form spleen colonies (day 0 equivalent) was measured by transplanting transduced cells after purification into lethally irradiated recipient mice and quantifying spleen colony formation 12 days after injection. Constitutive expression of FLT3-LM did not increase the CFU-S content compared to the GFP control. In contrast, AML1-ETO increased the CFU-S content 3.1 fold compared to the control (p < 0.002). Strikingly, co-expression of FLT3-LM together with AML1-ETO increased CFU-S numbers a further 2.1 fold for a net increase of 6.5 fold CFU-S compared to the control (p < 0.013) and thus demonstrating functional collaboration of these two genetic alterations in enhancing the CFU-S frequency. In an effort to characterize the domains responsible for the collaboration of the two aberrations, a FLT3-LM and a AML1-ETO mutant was generated: the FLT3-LM K672R mutant with loss of its kinase activity (FLT3-LM-KD) and the AML1-ETO mutant with a L148D point mutation in the Runx1 domain of AML1-ETO (AML1-ETO-L148D), previously reported to lack DNA binding activity. Expression of the constructs was tested by Western Blot or FACS analysis, in the case of the FLT3-LM-KD in addition for autophosporylation as a surrogate marker for kinase activity and for its capacity to induce IL-3 independent growth of infected Ba/F3 cells. Of note, the AML1-ETO-L148D was not able to collaborate with the FLT3-LM. Furthermore, the collaboration between AML1-ETO and FLT3-LM was dependent on its kinase activity as the FLT3-LM-KD did not collaborate with the fusion gene. Inhibition of the kinase activity of the FLT3-LM by the PKC412 inhibitor was tested in a delta-CFU-S assay after 48h incubation with the inhibitor: the compound induced a 62 % (42 versus 16 per 1x105 day 0 equivalent, respectively) reduction of the CFU-S frequency of cells co-transfected with FLT3-LM and AML1-ETO compared to the untreated control, whereas the CFU-S frequency of cells infected with the GFP control vector was unchanged by the inhibitor.
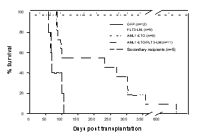
To further assess the potential collaboration of AML1-ETO with FLT3-LM, we carried out long-term BM transplantation studies using BM transduced with AML1-ETO or FLT3-LM alone or together. Over an observation period extending to 20.6 months no disease developed in recipients of BM singly transduced with AML1-ETO (3 x 105 to 4 x 105 highly purified GFP+ cells; n = 9) or FLT3-LM (7 x 104 to 2 x 105 highly purified YFP+ cells together with 3 x 105 – 1 x 106 non-transduced helper cells; n = 9). To obtain mice engrafted with AML1-ETO/FLT3-LM co-expressing bone marrow cells, mice were injected with a mixture of GFP+/YFP+ cells (range 1 x 103 to 5.5 x 104 cells) and non-transduced normal BM cells (range 2.3 x 105 to 1.9 x 106). All recipients of doubly transduced BM (n=11 from 5 independent experiments) succumbed to an aggressive acute leukemia after a median latency time of 233 days post transplantation (Figure 1).
At diagnosis the mice were moribund, cachectic, short of breath and suffered from splenomegaly (median spleen weight 441 mg). Peripheral blood and BM contained a high proportion of blasts and PB white blood cell counts were highly elevated in 5 of 11 animals with up to 430 x 106 cells/ml (range 2 – 430 x 106) compared to the GFP control (range 3.5-9 x 106) consistent with a diagnosis of acute leukemia. Additionally, mice were anemic with a 45 % reduction in erythrocyte counts compared to the mean count in the control. In 7 animals the morphology of the blasts was myeloblastic, whereas 4 animals were characterized by a lymphoblastic cell population. In two of the 7 animals with AML the blast population was accompanied by a dominant mast cell population with fine metachromatic granulation in the panoptic staining. The BM and spleen were infiltrated with up to 80 % and 80% blasts in the mice with AML and up to 85 % and 95 %, respectively, in the mice suffering from ALL .
In order to determine more precisely the immunophenotype of the leukemic population, flow cytometric analyses from bone marrow cells were performed. 7 animals suffered from AML with a Gr1/Mac1-positive cell population in the transduced compartment, which co-expressed Sca-1 (45,2 %, range 19 - 73). Of note, in 4 of the 7 animals with AML, co-expression of CD4 was detected in 21, 27, 31 and 32 % of BM cells. Three mice suffered from B-lymphoblastic leukemia with 90.4 % of the transduced cells expressing B220 (range 85 – 97) and lacking expression of myeloid antigens (Gr1+ 1.5 %, range 0.4-2.3; Mac1+ 1.7 %, range 1.4 – 1.9). One animal developed T-cell leukemia with co-expression of CD4 and CD8 (99 % CD8+, 86 % CD4+ in the transduced compartment) and expression of Sca-1 in 76 % of all cells. Histological tissue sections and immunohistochemistry were performed in two diseased mice with AML, including one of the animals with an increase in mast cells in the peripheral blood. Both animals showed multiple organ infiltration into hematopoietic and non-hematopoietic organs with effacement of the normal follicular architecture and the B220 positive B–cell population in the spleen and infiltration with blasts in the liver and spleen. Immunohistochemistry confirmed the diagnosis of AML: blasts were positive for myeloperoxidase but showed differentiation into more mature myeloid cells with positivity for chloracetatesterase (Fig.2).
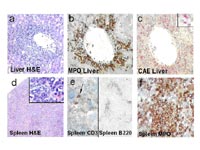
In the second case with AML, infiltration of organs with cells expressing mast cell specific tryptase and CD117 could be confirmed in the primarily and secondarily transplanted mouse indicating the presence of a malignant infiltrating mast cell population in this animal.
The leukemias were readily transplantable and with the same histomorphology within 106 days post transplantation (median survival 68 days, range 57-106 days; n=5)(Figure 1). Southern Blot analyses of bone marrow from leukemic mice revealed modest numbers of proviral integrations consistent with double infection and monoclonal or at most oligoclonal disease. Monoclonal or oligoclonal disease is consistent with the relatively small transplant doses used, but could also reflect a possible contribution of retroviral insertional mutagenesis to the transformation process. To further explore this latter possibility, 10 retroviral integration sites were subcloned and sequenced from 4 leukemic mice: all 10 sites were unique and thus there was no indication of a common integration site associated with the leukemic transformation. Moreover, 5 sites were intergenic or not linked to known genes. The remaining sites were in introns in a 5’ to 3’ orientation most likely to lead to gene knockdown rather than activation.
Outlook
Collectively, these data demontrate that the FLT3-LM is able to cause aggressive leukemia together with one of the most frequent fusion genes in AML, AML1-ETO, in transplanted mice. However, although the FLT3-LM occurs recurrently together with AML1-ETO, most of the patiens with FLT3-LM have a normal karyotype. The recent finding of the NPM mutation strongly associated with the FLT3-LM have pointet to the relevance of genetic aberrations not detectable by classic cytogenetics. Furthermore, results from cDNA microarray analysis revealed an aberrantly high expression of MEIS1 as well as HOXA9/A10 in AML patients with FLT3-LM. Thus, our project will now focus on the contribution of aberrant homeobox gene expression and NPM mutations in the pathogenesis of FLT3-LM positive AML.
Lit.: Schessl et al., The AML1-ETO Fusion Gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. Journal of Clin Invest. 2005 August 115 : 2159-68.


