Introduction
Leukemias are a model disease for cancer in general. Acute myeloid leukemia (AML) is a clonal disease originating from myeloid progenitor cells in the bone marrow. Unlimited expansion of a malignant transformed progenitor cell, the leukemia stem cell, and secretion of negatively regulating factors such as TGF-#, result in the clinical signs of acute leukemia: decreased normal progenitor cells with low platelet counts in the peripheral blood, anemia, and low granulocytic counts. Death of untreated patients most frequently results from sepsis or bleeding.
There is marked heterogeneity with regard to morphology and cell biology of AML. Although morphology is the hallmark of the diagnosis, it is itself of limited prognostic importance. Besides age, cytogenetic analysis is the most important prognosticator. Translocations such as t(8;21); t(15;17); or inversion inv(16) ) are associated with good prognosis (i.e. favourable karyotype), examples of a karyotype associated with poor risk include deletions of chromosomes 5 or 7, t(6;9), t(9;22), and others are associated with a standard risk, for instance a normal karyotype or trisomy 8.
Leukemias are frequently caused by aberrant transcription factors. For instance, all translocations associated with good prognosis, t(15;17), t(8;21), or inv(16), result in a fusion of two unrelated genes causally related to leukemogenesis. In the case of t(15;17), the two genes contribute to the fused leukemogenic gene are PML and RARa. This specific leukemia is characterized by a differentiation arrest at the stage of promyelocytes. The normal PML protein acts as coactivator of the well known tumor suppressor p53; it is a critical protein for all other ND10-associated proteins; it encodes a protein yielding in several pro-apoptotic signaling events. The fusion protein PML-RARa heterodimerizes with RXR, the retinoic X receptor, and binds to RA responsive elements in several RA target genes. PML-RARa then inhibits regular RA signaling in a dominant negative manner. Clinically, APL responds very well to a combined treatment of chemotherapy, and high doses of vitamin A, retinoic acid (RA). It is thought that the aberrant fusion protein PML-RARa causes a repression of RA inducible genes; very high doses of RA may overcome this repressive effect. Since the introduction of this specific biological therapy, ATRA in combination with anthracycline-based chemotherapy, cure rates have dramatically increased; in a recent review, Sanz et al. reported ongoing complete remissions as high as > 85% in adult APL patients (Sanz M.A. and Lo Coco F. 2005, Haematologica 90: 840-845). Introducing PML-RARa into suitable mice models, expression of PML-RARa itself is not sufficient to cause full leukemia. When co-expressing Flt-3- or ras-mutations, leukemias arise at high frequency. Interestingly, Flt-3 mutations are indeed very frequently found in APL.
AML with a particular poor prognosis (i.e. cure rates below 10%) are characterized by either specific translocations such as t(6;9), or a complex karyotype. In addition, AML carrying a deletion of the whole chromosome 7, -7, or del7q-, harbour a poor prognosis, the molecular background of which is poorly understood. Therefore, we performed gene expression profile analysis with AML characterized by -7 or del7q, and compared this to AML with normal karyotypes, as well as normal CD34 progenitor cells. A gene not previously associated with leukemias, the nuclear repressor Ski, was highly up-regulated in AML with -7 / del7q, as compared to normal stem cells and AML with normal karyotype. The current project was initiated in order i) to understand the specific role of Ski in AML with regard to biology, ii) to compare AML with Ski expression with regard to prognosis, and interaction with specific treatment strategies such as therapy with RA, and iii) to understand how Ski may enhance leukemogenesis.
Results
Analysis of leukemia and normal samples:
Expression profiling was used to discern the expression signatures between two different AML subgroups (AML with -7 / 7q- and AML with normal karyotype and normal Flt3) and normal bone marrow derived CD34 positive progenitor cells. 20 AML cases were selected out of 201 AML cases used in this study, nine bone marrow samples were from patients with -7 / 7q-, and eleven bone marrow samples were taken from AML with normal karyotype. In all cases, more than 60% blasts were counted at central analysis. Twenty three patients (mean age: 63 years; range: 46-82 years) undergoing total hip arthroplasty, due to coxarthrosis or fracture, were included in this study as control group for array analysis. Neither malignant nor chronic inflammatory or systemic diseases were present. The spongy bone tissue was carefully cut into small pieces, rinsed with saline and filtered. CD34+ selection of the bone marrow cell suspension was performed using magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). This procedure was repeated once. The mean purity of CD34+ cells after two sequential selections by magnetic microbeads was 76%. Each patient gave written informed consent to participate in the study. The ethics committee of the Philipps-University Marburg approved the study.
A cDNA chip consisting of 4.608 genes was used to derive expression profiles from nine -7 / del7q AML patients, which was first compared with CD34+ progenitor cells isolated from healthy human bone marrow (n=12). The statistical analysis revealed 70 genes discriminating between the two groups, with an adjusted p-value of p<0.05, of which 24 were up- and 46 down-regulated in the -7 / del7q AML samples. In a second set of experiments, AML with normal karyotype (n=11) were compared to normal CD34+ samples (n=11). In this experiment, 69 genes discriminated between the two samples at an adjusted p-value p<0.05, of which 18 genes were up-regulated, and 51 genes down-regulated in the AML samples.
Ski is up-regulated in many subtypes of AML, but not in AML CBF –AML and APL:
Among the genes most significantly up-regulated was the nuclear co-repressor Ski, which has been implicated in several different malignant diseases such as colorectal cancer and malignant melanomas. Ski is a negative regulator of signal transduction through TGF-# in vivo. Second, Ski is a cofactor for repression of the Mad family of transcriptional repressors. Third, Ski represses transcriptional activation by nuclear receptors, e.g. RAR#. When analysing a high number (183 cases) of primary AML cells using a real time PCR assay, we found Ski to be up-regulated most highly indeed in -7/7q- AML. In two groups, AML characterized by t(8;21) or inv(16) (i.e. CBF+AML), and in APL, characterized by t(15;17), Ski was not differentially up-regulated as compared to normal CD34 progenitor cells. Since Ski has been implicated in repressing nuclear co-receptors such as RARa, we figured that Ski may function as a repressor of RA signalling in AML, similar to the PML-RARa fusion gene in APL (not shown).
Ski blocks retinoic acid induced myeloid differentiation:
And indeed, expression of wild type Ski in the AML cell line, U937, blocked the differentiating efficacy of RA significantly, in contrast to a Ski mutant (L110P) unable to interact with N-CoR. In addition, luciferase assays showed that wild type Ski exerted this blockade by inhibiting retinoic acid response elements, and that this was not observed with Ski (L110P) (not shown).
These data suggested that blocking interaction of Ski with N-CoR and possibly histone deacytelases (HiDAC) might revert the differentiation arrest caused by Ski. We used the HiDAC inhibitor valproic acid (VPA) in these experiments. In additon, other HiDAC inhibitors were tested and proved to yield similar results (not shown). To this end, we took advantage of the promyelocytic cell line, HL60, which lacks PML-RARa expression. And indeed, incubation with RA and VPA alone was able to induce differentiation (Figure 1A: morphology; Figure 1B: CD11b expression), and this was more pronounced when both drugs were used simultaneously (Figures 1 A and B):
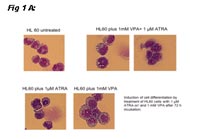

Thus, these data showed that myeloid HL60 cells are differentiated when they are incubated with RA alone or in combination with the HiDAC inhibitor VPA; the combination is more effective in this respect (read out CD11b expression). This is accompanied by a down-regulation of Ski. We also determined if Ski is more up-regulated in normal human hematopoietic precursor cells. CD34+/CD38- more un-differentiated cells expressed Ski more highly as compared to CD34+/CD38+ cells (not shown), arguing that Ski expression is associated with immature phenotype in hematopoiesis.
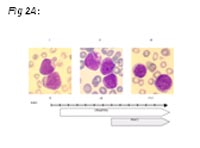
In order to ask if therapy with RA plus VPA would also be effective in human primary AML cells, we treated one individual suffering from AML at third relapse refractory to conventional treatment. As Figure 2 shows, cells were very immature at beginning of therapy, and showed signs of differentiation upon ongoing therapy (morphology; FACS analysis). Cytarabine therapy had to be started as cell counts increased during treatment.


Outlook
To find the cause for SKI up-regulation in -7 / del7q AML.
To study SKI expression in multiple AML samples in relapsed AML patients treated with RA and VPA.
To investigate whether Ski exerts its effects on retinoic acid signaling by the same set of genes as PML-RAR#.


