Introduction
The ultimate step towards understanding the human genome is the functional annotation of the 25.000 predicted genes and the countless number of proteins derived from them. In many species, insertional mutagenesis has been the key to functional genomics in the past. In the last 15 years, mutagenesis in the mouse has become the most relevant approach for extrapolation to human genetic disease because i) mouse and human embryonic development, anatomy and physiology are largely conserved; ii) 99% of mouse genes have direct orthologs in humans; iii) mouse genome structure and organization are closely related to those of the human genome and iv) the availability of the mouse genome sequences in combination with the embryonic stem (ES) cell technology allow the generation of specific and defined mutation in the mouse to model virtually any genetic human disease. Furthermore, by using high-throughput gene trapping and gene targeting approaches in ES cells systematic, large-scale genetic screens can be routinely performed. The gene trapping principle is based on the random integration of a reporter/selector cassette into the genome, resulting in the disruption of a gene. The reporter gene allows monitoring of gene expression and easy identification of the mutated locus.
Based on these developments, international programs such as the Knockout Mouse Project (KOMP) and the European Conditional Mouse Mutagenesis Project (EUCOMM) have been recently initiated with the aim to inactivate every single gene of the mouse genome in the context of an entire organism and to make these mutations freely available to the scientific community (1, 2). Based on an extensive analysis of the currently available gene trap resources, saturation mutagenesis is likely to be most effectively achieved by using a right balance between gene trapping and gene targeting. It is anticipated that gene trapping will be cost effective for mutating up to 50% of all genes in the genome after which trapping costs for novel genes will rise above those generated by gene targeting (8).
Results/Project Status
Genome-wide gene trapping program
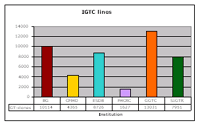
As yet, the German Gene Trap Consortium (GGTC) has produced 13,000 molecularly characterized mutant ES cell lines corresponding to 3,600 unique genes. Thus, contributing almost one-third of all gene trap lines to the public gene trap resource, the GGTC is one of the major players in the worldwide gene trapping effort represented by the International Gene Trap Consortium (IGTC) (see Fig. 2). The GGTC has recently added conditional features to its gene trap vectors that enable the induction of both null and spatially/temporally restricted mutations. In addition, the mutations the vectors create are multipurpose alleles amenable to a mutlitude of postinsertional modifications such as ectopic gene expression, protein tagging, cell ablation etc. Within the last year, the GGTC assembled the largest library of ES cell lines with conditional mutations in single genes, presently totalling over 1,000 unique genes (4). By using conditional gene trap strategy in the future, the GGTC will further increase the value of the international gene trap resource.
Based on these results and the extensive analysis of gene trap resources, the DHGP and NGFN-supported GGTC will be able to trap up to 50% of all mouse genes within the next five years in collaboration with its EUCOMM partners. In parallel, the GGTC will use European support, to inactivate untrappable genes (e.g. non-expressed or single exon genes) by homologous recombination using a generic gene trap style cassette.
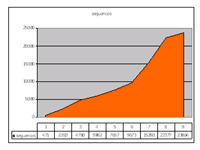
Figure 1 shows the overall performance of the GGTC. Owing to improved vector designs and sequencing strategies, as well as to extensive automation, gene trap line production has become significantly more efficient, particularly during the last two years.
All sequence tags from gene trap lines produced so far by the IGTC have been deposited in the NCBI genome sequence database (gss) and most of them have been annotated to ENSEMBL genes. More than 400 GGTC gene trap lines have been distributed to researchers worldwide in recent years with requests rising exponentially.
With the implementation of a dynamic vector controlling protocol which automatically monitors the trapping rate of each gene trap vector, the GGTC can now quickly switch to alternative vectors once the trapping rate with an individual vector becomes too low. Currently, the GGTC is using nine different retroviral gene trap vectors each designed for all three translational reading frames. Some of these vectors were designed to target signal peptide and/or transmembrane encoding genes.
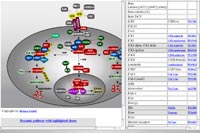
Finally, the GGTC has developed a dynamic signal transduction pathway tool which assigns gene trap mutations to individual components of a particular pathway. This will facilitate the genetic analysis of signal transduction pathways (Fig. 3).
Disease relevant mouse mutants
The GGTC has generated several mouse models of human disease, which have been phenotyped in part by the SMP's German Mouse Clinic (GMC). One example is a model for human nephrotic syndrome resulting from the disruption of the Nphs1 gene (6). The Nphs1 gene encodes Nephrin, which is a structural protein of the interpodocyte filtration slits and therefore crucial for the formation of primary urine. Mouse mutants develop characteristic features of proteinuric disease and die soon after birth.
A second example is a model for pulmonary emphysema and colon cancer resulting from the disruption of the gene encoding LTBP-4 (latent transforming growth factor beta (TGF-beta) binding protein 4). LTBP-4 is a structural component of connective tissue microfibrils and a local regulator of TGF-beta tissue deposition and activation (10, 5).
Finally, a third example is a model for a placental abnormality that leads to embryonic lethality. Mice with a disruption of the Bruce/Birc6 gene which is involved in the regulation of apoptosis die between day 11.5 and day 14.5 of embryonic development due to a lack of spongiotrophoblast cell proliferation in the developing placenta (4).
Outlook
The growing demand within the scientific community for mutant mice and therefore, mutant ES cell lines highlights the importance of the international gene trap resource. As a leading partner of the EUCOMM Consortium whose aim is the saturation mutagenesis of the mouse genome, the GGTC will contribute ES cell lines with conditional mutations in single genes induced by gene trapping and gene targeting. We anticipate that within five years a conditional allele for every single mouse gene will be available in ES cells. Once available, this comprehensive resource will form the basis for the establishment of mutant mouse strains and their phenotypic characterization.
Lit.: 1. Austin CP et al. The knockout mouse project. Nat Genet. 2004 Sep;36(9):921-4. 2. Auwerx J et al. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004 Sep;36(9):925-7. 3. Hansen, J. et al. A large scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA. 2003. 100, 9918-9922. 4. Hitz C et al. Progressive loss of the spongiotrophoblast layer of Birc6/Bruce mutants results in embryonic lethality. Genesis. 2005 Jun;42(2):91-103. 5. Koli, F. et al. Disruption of LTBP-4 function reduces TGF-beta activation and enhances BMP-4 signaling in the lung. J Cell Biol. 2004. 167, 123-133. 6. Rantanen, M et al. Nephrin TRAP mice lack slit diaphragms and show fibrotic glomeruli and cystic tubular lesions. J. Am. Soc. Nephrol. 2002. 13, 1586-1594. 7. Santti H et al. Disruption of the murine PIASx gene results in reduced testis weight. J Mol Endocrinol. 2005 Jun;34(3):645-54. 8. Skarnes WC et al. A public gene trap resource for mouse functional genomics. Nat Genet. 2004 Jun;36(6):543-4. 9. Schnütgen F et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci USA 2005. 102(20):7221-7226. 10. Sterner-Kock, A et al. Disruption of the Gene Encoding the Latent Transforming Growth Factor b Binding Protein 4 (LTBP-4) Causes Abnormal Lung Development, Cardiomyopathy and Colorectal Cancer. Genes Dev 2002 16, 2264-2273.


