Among the various strategies developed for the in vitro selection of recombinant antibodies, phage display has proven to be a robust and versatile technology and has yielded most of the recombinant antibodies generated so far. In the first decade of the development of antibody phage display, the technology has been driven primarily by the needs of therapeutics development. As a consequence, neither the process nor the antibody format have been optimised to yield reagents for research – last but not least for a price affordable for academic researchers (Fig.1).
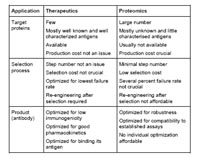
In the “postgenomic" era, antibodies against a large number of gene products from various genome projects are required, to be used in various immunological standard assays (Hust et al. 2004a). Current applications are mainly established for the use of mouse and rat monoclonal antibodies. Therefore, it is necessary to develop the antibody design in a novel way, so that recombinant antibodies obtained by phage display can be finally exchanged for animal-based IgG antibodies with no or minimal protocol change. Another task of this
subproject is work towards a “minimal step” protocol, as throughput per cost of the phage selection pipeline was a minor issue in the development of therapeutical reagents. Many mouse monoclonal immunoglobulins (IgG) with poor monovalent affinities are useful research tools due to the cooperativity of both antigen binding sites. To imitate this feature, a dimerisation tag will be utilised to combine two monovalent recombinant antibody fragments yielding an avidity effect similar to IgG antibodies.
Project Status
Antibody formats and vectors for phage display Various leader sequences, promoters, tags and cloning strategies have been successfully used in phage display vectors, with none of them providing a perfect construct for the needs of proteomic research. In conclusion, there are many different ways to succeed with various factors that can be altered depending on individual applications. Two factors seem to be most important – first the careful control of phage biology during the panning process and second the use of an antibody phage library as large as possible in respect of its genetic complexity.
Therefore, the first goal of this subproject is to optimise the antibody format and phage vector design prior to the
construction of new scFv and Fab libraries of a diversity sufficient for the high throughput generation of research antibodies. It is possible to optimise both the antibody format and the phage vector design, but systematic studies are rare (Hust et al. 2004b). In the SMP Antibody Factory, here is a main focus set on the Fab format, resulting from its better stability and its compatibility to existing immunological applications, when compared to scFv fragments. However, due to the need to assemble a Fab molecule from two
different polypeptide chains, a well balanced phage display and bacterial expression system is required. Various Fab phage vector designs have been constructed and were compared in regard to antibody display on the phage and soluble Fab expression in E. coli. Additionally, monovalent and multivalent display of Fab fragments was compared using the helper phages VCSM13 and Hyperphage,
respectively. The results suggest to use a bicistronic vector design with a fd-fragment–pIII fusion and an arrangement of the fusion cassette at the 3’ end of the operon for the construction of future Fab phage display libraries (Kirsch et al., 2005).
Furthermore, the insertion of an amber stop codon between fd-Fragment and pIII allows both phage display using SupE44-E. coli strains and soluble Fab expression with no need of subcloning of selected antibodies into another expression vector. The resulting pHAL phage vector family will be continuously upgraded to finally combine all features examined in this subproject.
Minimal step protocol
Another important task of this subproject is to reduce number of steps for phage display and the subsequent analysis to enhance the throughput of antibody selections in the SMP “Antibody Factory”. Therefore, different antibody phage libraries were tested to optimise the panning protocol and the screening of antigen specific binders. Adapting these methods to a automatted workflow is done in cooperation with the two SMP partners at RZPD and MPI.
Panning against peptide antigens
In proteome research, it is very frequent that protein antigens are not available and cannot be expressed. Here, peptides representing linear epitopes on the surface of the protein could be an alternative antigen for the selection of antibodies. Surface accessible peptides are calculated and synthesized by the SMP partner “Peptide technology”. Short linear peptides are much more flexible than peptide epitopes
enclosed in the protein framework. Therefore, they do not exactly represent the three-dimensional structure within the protein. Phage display and animal immunisations using linear peptides as antigen so far often resulted in antibodies not recognizing the native protein. Using a larger set of different peptides from the same protein may increase the probability to get antibodies binding to the native antigen.
Additionally, posttranslational protein modifications, which are difficult to isolate as native antigen, can be substituted by chemically modified peptides.
Panning on array
Another work package deals with the establishment of a polyclonal array panning method for antibody display libraries in close colaboration with the subproject “Peptide Technology”. The principle is based on the use of an array of up to 2000 peptide antigens directly synthesised on a chemical inert support (polypropylene) and promises to minimise unspecific antibody phage binding and to allow
more stringent selection conditions. The intended benefits concern both improved throughput, and reduction of the library volume required per antigen by at least an order of magnitude.
Bivalent recombinant antibodies
Further work is done on the definition of an optimal tag allowing convenient dimerisation of antibody fragments derived from phage display libraries to utilise the “avidity” effect of two antigen binding sites. Moreover, this tag shall be useful for an ordered surface attachment of antibody fragments, e.g. on microarrays. In addition, it should also enable the preparation of a high quality antibody fraction
(“ready to use”) by affinity chromatography from crude E. coli culture preparations. Further advantages shall be achieved by the dimerisation tag to improve the compatibility of recombinant antibodies to mouse IgGs and standard immunoassays as shown in Fig. 2. Several potential useful dimerisation tags have been fused to test scFv fragments and are currently under investigation.
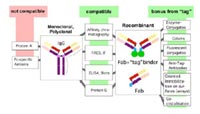
Proof of application
The final goal of the SMP “Antibody Factory” is the generation of recombinant antibodies which can be applied for immunoassays commonly used in research and clinical diagnostics. Therefore, scFv and Fab fragments obtained in the SMP have to be tested in immunological standard assays (e.g. immunoblot, immune fluorescence flow cytometry etc). The performance of the phage antibody formats will be compared in respect of signal to signal intensity, noise ratio and specificity, to “conventional” antibody molecules by
utilising recombinant IgG with the same variable region produced in eukaryotic cell culture.
Outlook
Almost every parameter of the complex pipeline of the in vitro antibody selection has to be adapted to the needs of least effort and higher throughput. Further, the antigen preparation has to be integrated into this process, and the validation of specificity and affinity of the obtained recombinant antibodies has to be parallelised as well. As a result, new methods have been developed. Novel antibody
formats currently under examination may also flow into this process. This subproject also aims to establish a “polyclonal panning” format on peptide arrays which could dramatically accelerate the throughput of antibody generation with reduced library consumption and increased specificity. It will further combine the benefits of a polyclonal recognition with the reproducibility of a monoclonal antibody preparation.
In summary, this subproject in closely interconnection with the other three projects of the SMP “Antibody Factory” is aimed to lay the foundation for an integrated structure for theproduction of antibodies for proteome research.
Lit.: 1. Hust, M. and Dübel, S. Mating antibody phage display to proteomics. Trends in Biotechnol. 2004 Jan;22(1):8-14. 2. Hust, M. and Dübel, S. Phage display vectors for the in vitro generation of human antibody fragments. Methods Mol. Biol. 2004;295:71-95. 3. Kirsch, M., Zaman, M., Meier, D., Dübel, S. and Hust, M. Parameters affecting the display of antibodies on phage. J Immunol Methods 2005 Jun;301(1-2):173-85.


