The functional annotation and characterisation of proteincoding gene products derived from the human genome sequencing initiative is one of the biggest challenges we currently face in molecular biology. In the light of this demanding task binder molecules, such as antibodies, fulfil an important role, since they are currently the best tools for the detection of the expression, the localisation and the site
of function of proteins and their complexes in the context of a cell or organism. Their application in systematic analyses of the proteome will take us a step closer in the understanding of the functioning of a cell and provide information on the link between the sequence deposited in the genome and the function exerted by the protein. Ultimately, this work demands large sets of specific binders as research tools and although many thousands of antibodies are commercially available, to many proteins still no binders are on hand (Konthur et al., 2005).
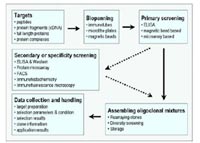
The overall aim of the SMP “Antibody Factory” is the development of streamline technological tools and to implement processes that facilitate the generation of recombinant antibody molecules by phage display in a highthroughput and systematic fashion against human gene products for the use in proteomic research. Within the SMP “Antibody Factory”, our subproject is concerned with the automation of panning and evaluation procedures and is dedicated to the assembly of a selection pipeline resulting in oligoclonal mixtures (pools of different antibodies directed towards the same target protein) of phage display derived antibody fragments.
Project Status
The pipeline consists of individual modules concerned with different tasks necessary in a production line addressing topics such as source of target, target expression, semiautomated selection, primary and specificity evaluation, assembly of oligoclonal mixtures and data handling. Due to its open architecture, the pipeline can be easily modified and expanded, and shows general applicability to any other type of in vitro selection schemes next to phage display (Fig. 1). Starting from this, four work packages have been formulated.
Expression & purification of antigen targets
For the successful selection of antibodies by phage display, solubility, homogeneous folding and presentation of the antigen are important issues. Yet, the generation of soluble recombinant target protein is currently still one of the major bottlenecks. Within this work package, we are primarily focusing on the expression of recombinant proteins using E. coli as an expression host. Currently, we are investigating
the possibilities to further streamline the available E. coli protein expression protocols by supplementing different solubility-increasing additives to the medium. Another key issue of protein expression for our purposes of semi-automated selection is the expression and purification of equal amounts, as well as the biotinylation of the proteins. Therefore, we are currently investigating the use of in vitro
transcription/translation-coupled protein expression systems in collaboration with the RiNA GmbH in Berlin - a partner within the NGFN-2 programme. Furthermore, we are planning to set up a streamlined single-step purification protocol, which allow the coating of the magnetic particles directly from cell lysates.
Generation of a new antibody library
As a direct consequence of the growing demand of research antibodies in the field of functional genomics and proteomics necessary for evaluation purposes, in this work package we will generate a novel library that yields antibody fragments that can directly substitute full IgG and polyclonal antibody sera in commonly used experimental techniques. The most important features of the library format we are planning for includes ease of handling, robustness and yield of antibody production. For this reason, we are designing the new
antibody library in the Fab format and the library will be based on a single antibody framework with directed diversification of selected amino acid positions in the complementarity determining regions (CDR) of the heavy and the light chains.
Another key issue for the new library is the rate of presentation of the antibody fragment on the phage particle. From our experience in phage display automation it has become evident that high percentage of functional antibody presentation is of crucial importance for efficient robotic selection.
Automated selection of antibody fragments
We have set up a method for selecting specifically binding single chain antibody fragments (scFv) from phage display libraries in a semi-automated fashion utilising a magnetic bead-handling robot in 96-well plate format (Konthur & Walter, 2002). During the process, specifically binding antibody fragments are enriched on target molecules - tagbound to magnetic beads. We were able to demonstrate this
working principle on different types of molecules, such as recombinant and natural proteins, peptides and small chemical compounds (Rhyner et al., 2003). In this work package, we will further improve the selection steps to enable higher throughput.
Another aspect of selection improvement is the modification of the screening protocol to obtain antibodies recognising different types of epitopes, such as linear and conformational epitopes, as well as antibodies directed towards posttranslational modifications of the target molecules. These antibody fragments are of particular interest for proteomic research, but also for in vivo diagnostic and therapeutic
applications.
Downstream evaluation of selected antibodies
Automating the selection process on its own largely increases the throughput of antibodies that can be generated, but also shifts the bottleneck of the selection pipeline further towards the isolation and evaluation of monospecific binders. Therefore, we are developing and improving different techniques for downstream characterisation of selected antibody fragments for both steps of the pipeline, the primary and secondary screening (Fig. 1). The techniques we are currently working on include enzyme-linked immunosorbent assays (ELISA) in microtitre plate format and on magnetic beads, and more recently the

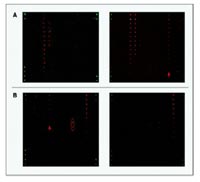
use of protein microarrays. Protein microarrays can be used at multiple steps within the pipeline. For example, we use protein microarrays for the monitoring of selection rounds in respect of increasing specificity (Fig. 3). Similarly, we are using protein microarrays for the comparison of monospecificity and sensitivity of individual antibodies or polyclonal mixtures, and compare these with commercially available antibodies (Fig. 2). Currently, we are in the process of further advancing the use of protein microarray
applications by combining the described features with the multiple spotting technique (MIST; Angenendt et al., 2004), which will allow the parallel testing of thousands of single antibody-expressing clones at a time on a single chip in a multiplex assay format.
Outlook
Within our subproject in the SMP “Antibody Factory”, the major task is the linkage of available techniques to create a streamlined pipeline for the generation and evaluation of antibody fragments. However, all the techniques currently under investigation, such as protein expression & purification in microtitre plate format, selection of libraries on magnetic bead-handling robots, protein macro- and microarrays for specificity evaluation, and the application of MIST (Multiple Spotting Technique) for the evaluation of hundreds of
monoclonal entities in parallel, still need considerable amount of adaptation to our needs and fine-tuning. We envisage, however, that by the end of the funding period the successful implementation of all modules into the described pipeline will yield a system capable of generating oligo- and monoclonal antibody fragments against ~500 target proteins per year.
Lit.: 1. Konthur Z et al. Perspectives for systematic in vitro antibody generation.Gene. 2005; in press. 2. Konthur Z and Walter G. Automation of phage display for high-throughput antibody development. DTT:TARGETS 2002;1:30-6. 3. Rhyner et al. High-throughput isolation of recombinant antibodies against recombinant allergens, Biotechniques 2003;35:672-4. 4. Angenendt et al. Seeing better through a MIST: Evaluation of monoclonal recombinant antibody fragments on microarrays. Analytical Chemistry 2004;76:2916-21.


