Introduction
A new technology addressing the toponome
The hierarchy of cell function comprises at least four distinct functional levels: genome, transcriptome, proteome, and toponome. The toponome (rule of protein arrangement) is the entirety of proteins, protein-complexes, and protein networks traced out on the single cell level in the natural environment of cells in situ (e.g. tissues). Whole cell protein fingerprinting (WCPF/MELK) enables the quantitative mathematical description and exploration of the toponome by using large dye-conjugated tag libraries (antibody, or any affinity reagents) co-localizing proteins (tens to hundreds simultaneously) by running iterative rounds of protein-tagging, imaging and bleaching on one fixed biological sample (Lit 1). Thereby the technology addresses the fact that each protein must be at the right time at the right place at the right concentration in a cell to interact with other proteins assembled in a spatially organized network. The entirety of these toponome units represents the total functional code, or plan, of a cell (Lit 2-5). The complete toponome is therefore as fundamental a data set as the genome or the proteome. Earlier studies on cell types other than neurons have demonstrated the principle feasibility of the MELK technology (Fig. 1).
The biological problem: Local rules of synaptic protein networks
All thoughts and actions are encoded in patterns of neuronal electrical activity. Circuits of nerve cells connected by synapses are dedicated to processing information in these patterns.
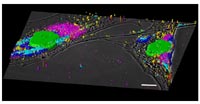
Information is not only transmitted across the synapse but also monitored by postsynaptic molecular machines. These machines are macromolecular complexes of approximately 100 different proteins organized into a network of protein interactions. These networks have never been seen directly, which is a major obstacle for describing combinatorial protein arrangements encoding local functionalities in the brain. Importantly, a detailed local quantitation of synaptic multi protein clusters and their network properties will shed new light on many human degenerative diseases, such as Alzheimers, Parkinson, and others.
Goals
This project intends to analyze by using the MELK robot technology the organization of the cell surface toponome of neuronal synapses in situ. The special focus will be the analysis of nerve cells in the brain of mouse models of neurodegeneration provided by the consortium. On the basis of preliminary observations we wish to examine the hypothesis that individual nerve cells in defined neurodegenerative disorders display abnormal protein clusters on the cell surface and in postsynaptic structures which are basic modules of abnormal signaling cascades and therefore highly relevant for new therapeutic avenues. The resulting new systems biology tool, providing a map of the synaptic toponome in health and disease, will be the basis for multiple cooperation’s within the consortium.
Project Status
On December 15, 2004, the consortium has decided to study multiple mouse models of neurodegenerative disorders. Following this decision, we had to adapt our project related to an appropriate measuring capacity of MELK robots and software tools. This task required the employment of specialists in the field of engineering and informatics in our project. Since the start of the project on April 1, 2005, we are following our milestone plan in two respects: (i) adapting the measuring capacity of MELK robots by extending the throughput to be able to map the synaptic toponome of at least 3 different mouse models; (ii) calibrating an antibody library to mouse brain tissue sections. We are presently working on both topics and have already succeeded in automating the 3D imaging procedure of synaptic brain regions for more than 20 proteins simultaneously as well as extending the measuring capacity of one MELK robot by factor four. Furthermore we have succeeded in defining a tag library recognizing 19 synapse-associated proteins at once in one single mouse brain tissue section in multiple locations.
Outlook
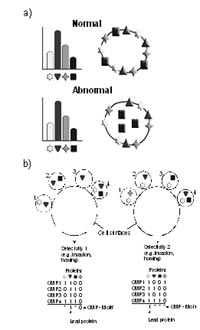
The next step will include a further extension of: (a) the MELK capacity, and (b) the number of synaptic proteins to be co-localized simultaneously. We expect to be able to measure systematically mouse brain tissue sections in the course of the next 2 months, thereby finalizing the technology part and starting the biological part of the project, probing the theory of toponome (Fig. 2) in synapses of the brain.
Lit.: 1. Schubert W. Automated determining and measuring device and method. USA patent 6,150,173. 2. Schubert W. Topological proteomics, toponomics, MELK technology. Adv. Biochem. Engin. Biotechnol. 2003; 83: 189-209. 3. Schubert W. Exploring molecular networks directly in the cell. Cytometry (in press). 4. Schubert W. Cytomics in characterizing toponomes: breaking the biological code. Cytometry (in press). 5. Schubert W, Friedenberger M, Bode M. Molecular networks in morphologically intact cells and tissue – challenge for biology and drug development. In: Hamacher M, Marcus K, Stühler K, van Hall A, Warscheid B, Meyer HE (eds). “Proteomics in drug research”. VHC-Wiley, (in press)


