Introduction
MicroDiscovery is a leading provider for high quality bioinformatics solutions in the areas of innovative diagnostics, personalised medicine and bio-molecular research. We develop customer oriented software solutions, successfully employed as certified components for in vitro diagnostic systems and other medical applications. One of the core competences of the company is the consequent quality assurance of our products in consideration of all relevant directives and standards (ISO 9001, ISO 9000-3, ISO 13485, 93/42/EWG, 98/79/EG). MicroDiscovery offers its customers and partners bioinformatics products developed according to industrial standards and services for the highest quality requirements. Our flexible and performance-oriented bioinformatics unit provides a wide range of professional services for our customers - fast, reliable and cost-effective. The youngest business unit is dedicated to the emerging field of systems biologiy.
Biological experiments have traditionally been carried out in vivo or in vitro but recent advances in biomedical research techniques open up a new direction: in silico modelling and simulation of biological processes (Fig. 1). Several projects are proceeding along these lines including academic research (e.g. E-CELL by Tomita et al., and simulations of biochemical pathways by Palsson et al.) and industrial efforts generating models for cellular metabolism (Genomica), dynamic disease simulation (Entelos) and models of ion channel function and cell signalling (Physiome).
Understanding complex, polygenic diseases can only be achieved by integrating data from the different functional genomics experiments (gene and protein expression, sequence data, etc.) with medical, physiological data and information on environmental factors. The aim of this project is to develop a knowledge, integration and modelling platform for the in silico representation of neuro-degenerative diseases. To this goal a number of more specific partial tasks has been identified that will be addressed within our project:
- Disease Classification: Analysis of the data with the specific goal of finding and evaluating different combinations of biomarkers for their efficiency in classification and prediction of neuro-degenerative disorders notably in an early stage of disease.
- Disease Modelling: Formulation of a generalized disease model and understanding of the time course of pathogenesis by means of mathematical modelling for neuro-degenerative diseases.
- BrainProfileDB: A data base for storing and accessing the data generated in the core studies of this project. Different experimental profiling techniques will be represented within a uniform framework.
- KinetXbase A specialized platform for mining and visualisation of different types of protein interaction data (e.g. BiaCore, Fluorescence Polarisation).
Project Status
Disease Classification and Prediction
This project is proceeding in close cooperation with the clinically oriented groups (Wiltfang, Kretzschmar, Schubert). Principal goal is the identification of a suitable set of biomarkers for the classification and prediction of neuro-degenerative diseases.

Emphasis is put on the application of clustering analysis for the identification of new hitherto unknown or insufficiently classified subtypes of neuro-degenerative diseases (Fig.2). Current efforts are aiming at the establishment of new methods for the selection of optimised biomarkers for diagnostic purposes. For this purpose improved methods for the treatment of unknown classes were developed and tested. Starting from the validated data set constructed together with the project partners MicroDiscovery will employ a set of established and newly developed statistical methods for definition and test of a suitable set of biomarkers.
Disease Modelling
Disease modelling will be performed based on the data gathered from different project partners and stored in the BrainProfileDB. Documents for experimental planning of the core studies in mice models within the SMP-Proteomics consortium are worked out in cooperation with the project partners to guarantee a accurate experimental design for the entire study. A web based system for the project-wide knowledge-management was set up, aim of this project is to establish a common platform for storing knowledge on neuro-degenerative diseases. Different platforms for modelling biological systems are being tested with respect to their suitability for the purpose of modelling neuro-degenerative disease processes.
BrainProfileDB
BrainProfileDB is an interactive data management system specifically tailored to the requirements of the Brain Proteome Project. The specific needs evolve from the fact, that a complicated disease process is investigated in several different mouse models employing different techniques and measuring numerous molecular parameters (Fig. 3). The data base and interface design are developed for the user friendly accessibility of the large data sets having substantial internal structure hiding most of the complexity of underlying queries at the same time.
In order to build on the know-how acquired in previous related projects an substantial effort was dedicated to the determination of the scientific requirements of the system. For a system employed in a scientific environment precise and consistent requests of the partners involved are difficult to acquire, therefore an iterative approach was chosen. Up to now MicroDiscovery developed two prototypes of the database system. These prototypes were implemented, tested and evaluated together with the project partners.
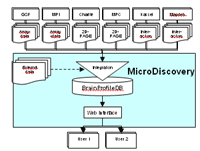
The resulting system is composed of several interacting sub-systems and is presented in Fig.3 An initial collection of experimental techniques mapped comprises biochips from several platforms: gene expression chips Affymetrix, customized glass slides from Prof. Beckers (PPO-S31T01) and Prof. Pääbo (PPO-S25T11) and validated results from 2D-gels (Prof. Klose, PPO-S02T03). Planned extensions in a later phase of the project will include protein interaction data (Prof. Herberg, PPO-S22T02). To achieve an efficient integration, a suitable interface to KinetXbase described below will be established. MALDI data (Prof. Meyer, PPO-S03T05) will be included with an interface to the Proteomscape data base.
KinetXbase
Following and extending MicroDiscovery's previous development KinetXpert in collaboration with our partner Prof. Herberg (PPO-S22T02) a platform for storing and accessing multiple types of interaction data will be designed and developed. This data management system is capable to represent different types of molecular interaction data in terms of a set of common properties including association and dissociation constants. The data types supported in the first release of the system are: Fluorescence Polarisation, Surface Plasmon Resonance, Stopped Flow, Isothermal Titration Calometrie.
Outlook
Data analysis and integration are a central and challenging task for a large and heterogeneous consortium like the Brain Profile Project. MicroDiscovery develops a sophisticated IT-infrastructure for the Human Brain consortium not only enabling the data exchange of the partners but enabling the user friendly data query and retrieval for all data sources defined as relevant. For guarantying the data quality necessary SOPs for data import and specific interfaces with appropriate filters will be provided. The planned system is unprecedented in respect to data types to be integrated and quality standards realised. The design of the system is kept flexible enabling the adaptation to new data types or changing user requirements. The data management system described is an excellent example for the sustainable achievement the cooperation of academia and SMEs are able to generate.
Lit. 1. Tomita M. Whole-cell simulation: a grand challenge of the 21st century. Trends Biotechnol. 2001 Jun;19(6):205-10. Review 2. Palsson B. The challenges of in silico biology. Nat Biotechnol. 2000 Nov;18(11):1147-50. 3: Challapalli KK, Zabel C, Schuchhardt J, Kaindl AM, Klose J, Herzel H. High reproducibility of large-gel two-dimensional electrophoresis. Electrophoresis. 2004 Sep;25(17):3040-7. 4. Pelludat C, Prager R, Tschape H, Rabsch W, Schuchhardt J, Hardt WD. Pilot Study To Evaluate Microarray Hybridization as a Tool for Salmonella enterica Serovar Typhimurium Strain Differentiation. J Clin Microbiol. 2005 Aug;43(8):4092-106 5. Nils Blüthgen, Karsten Brand, Branka Cajavec, Maciej Swat, Hanspeter Herzel, Dieter Beule Genome Informatics, 2005 Aug, Volume 16, Nr 1, 106-115.


