
Introduction
Substantial bioinformatics and data analysis expertise that works close together with the experimental projects is key to their mutual success, and thus a prerequisite for the progress of SMP-Cell. Functional genomics experiments generate huge amounts of data that need to be processed, stored and statistically analysed (1). Results have to be presented to the scientists, and connected with other relevant in-house or external information for evaluation and mining. Finally, this integrated information has to made accessible to the scientific community.
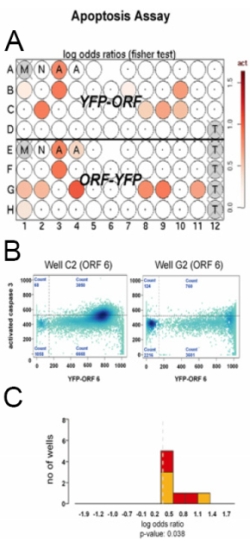 | |
| Fig 1: Statistical analysis of primary data. Several plots are produced for visualisation and quality assessment. A: Plate plots as overview for 96 well plates. B: Modelling of cellular effects. The scatterplots show the two fluorescence parameters from FACS data for protein abundance and effect size respectively C: Meta analysis of multiple experiments where significant deviations in measurements from a control group are detected. | |
Project Status
Protein annotation and analysis
All experimentally investigated proteins automatically undergo an exhaustive bioinformatic analysis (3). This comprises similarity searches, protein domain architecture determination, and prediction of physicochemical characteristics and secondary structure, using a wide variety of methods in combination with the most up-to-date public databases. This analysis is performed utilizing the HUSAR-system (4), and the resulting data are stored into an analysis database. The results are updated regularly, for instance BLASTP2 similarity searches are calculated on an weekly basis.
Statistics on primary experimental data
We have put special emphasis on the development of visualisation tools (Fig. 1) that quickly enable us to assess different aspects of the data, e.g. its quality, and to understand in necessary detail how the automatically-generated results were computed.
To model and detect the effect of a perturbation (e.g. over- or under-expression of the protein encoded by the gene/ORF of interest) on the cellular system, we use a range of methods from statistical regression analysis (linear models and their generalisations) to hypothesis testing and mixture modelling approaches. The general statistical models include three components: the biological effect of interest (e.g. the induction of chemo-resistance), systematic background effects (e.g. instrument drifts), and stochastic effects (biological and measurement noise).
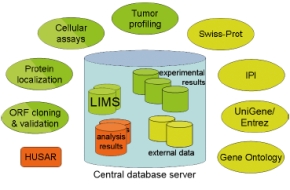 | |
| Fig 2: Data from several sources are integrated and stored on a central database server, facilitating data mining and analysis. Green: experimental data, orange: data from bioinformatics protein analysis, yellow: data from external publicly available databases. | |
Data integration and mining
All generated data (experimental results, bioinformatics data) and data from selected publicly available sources (6-8) is stored in databases on a central database server (Fig. 2).
The information is queryable and comprises:
• cDNA-annotation data
• experimental results from protein localisation and functional assays
• results from the automated bioinformatic protein analyses
• data from NCBI (Gene, UniGene), EBI (IPI, GO) and SIB (Swiss-Prot)
The data from the external sources is loaded by specialized applications and updated when new datasets become available.e have designed a statistical software package for the analysis and presentation
Outlook
The currently established databases offer a rapidly growing wealth of information on a large number of proteins. These will be extended to accommodate further experimental results from ongoing assays, e.g. using RNA interference technology. Together with cooperation partners, we will extend the automatic bioinformatic protein analysis by the development of new analysis procedures with emphasis on domain annotation/identification and secondary structure prediction. Further external databases will be integrated to allow for a continuous enhancement of data mining and to provide the experimental projects of SMP-Cell with most comprehensive information on the genes and proteins, and on their relevance in disease.
Lit.: 1. Wiemann S et al. From ORFeome to biology: a functional genomics pipeline. Genome Res. 2004 Oct;14(10B):2136-44. 2. Bannasch D et al. LIFEdb: A database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system. Nucleic Acids Res. 2004 32(1):D505-8. 3. del Val C et al. High-throughput protein analysis integrating bioinformatics and experimental assays. Nucleic Acids Res. 2004 32(2):742-8. 4. Ernst P et al. A task framework for the web interface W2H. Bioinformatics. 2003 Jan 22;19(2):278-82. 5. Arlt D et al. Functional profiling: from microarrays via cell-based assays to novel tumor relevant modulators of the cell cycle. Cancer Res. 2005 65(17):in press. 6. Boeckmann B et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003 Jan 1;31(1):365-70. 7. Maglott D et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005 Jan 1;33:D54-8. 8. Rodriguez-Tome P. EBI databases and services. Mol Biotechnol. 2001 Jul;18(3):199-212. 9. Kelso J et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003 Jun;13(6A):1222-30.
Please visit the Project's Website


