
Introduction
Quality management and standardization (QM&S) have become increasingly important issues in biomedical research. The quality of the material has substantial impact especially when clones and other molecular biological material is exchanged between groups and is fed into high-throughput experiments. The exchange and comparability of data and information strictly depends on common standards that need to be established and adopted in the production processes of resources and in their exploitation in experimentation. QM&S is relevant both from the labor and cost perspectives, as tested and standardized protocols increase efficiency when different labs contribute to the same resource, or when different labs use the same resource and experimental results are to be compared. The SMP-Cell is a highly interactive NGFN platform where substantial resources are generated and utilized by the different partners. Activities of QM&S within SMP-Cell are thus structured according to our main goals (Fig. 1): i) Quality control of molecular biological resources ii) Establishment and optimization of standardized protocols iii) Contribution to the QM&S initiatives within NGFN and beyond in order to establish and put forward common standards that shall increase the quality and reliability of data and the comparability of deduced information.
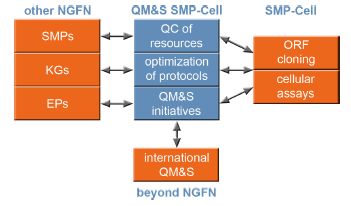 | |
| Fig 1:Main project activities of QM&S within SMP-Cell. Quality-controlled material is mainly Gateway entry clones and siRNAs; protocol optimization at present is concentrated on high-fidelity PCR in ORF cloning and on quantitative RT-PCR for QC in RNAi; contribution to QM&S initiatives within and outside NGFN is in the form of defining criteria for quality and annotation of ORF clones and comprehensive description of functional assay in terms of experimental conditions and standardization of data output formats. | |
Quality control of resources cDNA clones and siRNAs constitute the two major resources that are shared between partners within SMP-Cell and with partners of the NGFN. The clones are generated within the SMP-Cell project “The German cDNA Consortium: Cloning of ORF Resources”, mostly via RT-PCR or utilizing the full-length cDNA resources of the German cDNA Consortium that were generated within DHGP and NGFN-1 (1, 2). The establishment of standardized protocols and a strict quality control of products had been installed within the consortium to generate a high-quality cDNA resource . A similar standards for quality control of cloned material is mandatory to be continued in SMP-Cell, before the ORF resources can be exploited in cellular functional gene analysis that is carried out again at several partners within the SMP. Open reading frames (ORFs) are cloned into Invitrogen’s Gateway cloning system (3), and sequence validated as one most important step in QC. Thus far we have sequenced some 1,200 entry clones, another 1,500 ORF clones will be generated and sequence validated within NGFN-2. Only entry clones that have passed QC are used for shuttling into expression vectors and feeding into cellular assays (4). A variety of expression vectors has been mostly generated within SMP-Cell, the expression constructs are used throughout the SMP and by collaboration partners in SMPs and KGs. siRNAs are used within the SMP-Cell project “Cancer-relevant Cellular Assays” and by NGFN partners from disease-oriented networks. We tested enzymatically-generated esiRNAs and chemically-generated siRNAs from three different suppliers for their functionality in three different cell lines. Quality of knock-down was measured via qRT-PCR of the target genes/mRNAs. Cell-type and concentration dependent effects and preferences were observed with the different siRNA resources. RNAi appears to be an extremely valuable technology, however there seem to be some challenges that require further work to be carried out in order to understand the mechanisms and principles.
Optimization of protocols - SOPs
Standardized protocols require continuous optimization to match the often changing needs within cooperative projects and to take reference to improved technical knowledge, tools, and methodologies. Our current optimization activities concentrate on the cloning of ORFs and on quantitative RT-PCR (qRT-PCR). The SMP-Cell QC&S project supports the experimental projects and partners with expertise and testing capacities towards the establishment of common SOPs (Fig. 2).The fidelity of the DNA polymerase is a key parameter for the successful cloning of intact ORFs. PCR-errors are among the major obstacles within the ORF cloning process and continuous effort is invested to keep their numbers low. With a growing number of high-fidelity DNA polymerases available, we conducted an evaluation of the fidelity and processivity of different such enzymes. We tested 20 high-fidelity DNA polymerases and polymerase mixes using the LacI assay (5). The results of this test were used to optimize the fidelity of the PCR step in our cloning procedure, while maintaining reasonable yield and cost as was required in our high-throughput experimental set-up. Quantitative RT-PCR is a most relevant means of quality control within the functional assays of SMP-Cell, and used for instance to validate RNAi-induced gene knock-down. However, this fast and well-established method is still expensive when performed at a large scale. We are currently evaluating the reproducibility of the Exiqon qRT-PCR probe set, which covers most known genes with only 90 probes, and is a potentially cost-effective alternative to standard Taqman analysis
QM&S initiatives
A consensus on standards has to be reached between the participating groups and projects as a prerequisite for the development of standardized nomenclatures, molecular biological material, and protocols. Common quality criteria for material need to be established and SOPs have to be worked out and adopted. We actively participate in this process within the NGFN and beyond (Fig. 3), as the tight quality control of processes and material is one of the most relevant issues in SMP-Cell, where the production and exploitation of resources is conducted at different locations. The continuous tracking of material and data needs to be realized for the collection of relevant and reproducible data and information.
Within the NGFN, we have defined precise criteria for the annotation and quality control of ORF entry clones, that have to be met before any clone is released to enter the experimental pipeline in cellular functional gene analysis. An ORF entry clone must be verified to contain the expected ORF, that ORF must be in the correct reading frame relative to the cloning site, it must not contain any frameshiftmutation, and any silent or amino acid exchange mutation need to be clearly annotated. Current standardized protocols and developed standards on nomenclature and plasmid expression vectors are disseminated via the QM&S pages of the SMP-Cell homepage at http://www.smp-cell.org/ groups.asp?siteID=17, and with NGFN partners via the QM-page of the NGFN internal homepage. A discussion forum has been implemented in the SMP-Cell intranet pages, where relevant issues on SOPs and quality control are posted and discussed. In contrast to molecular cloning, QM&S for high-throughput cellular assays is still in its infancy. Within the first funding period of NGFN, several cellular assays were developed that monitor cancer-relevant biological processes (see SMP-Cell project “Cancer-relevant Cellular Assays”) (4, 6). Furthermore, the same genes and proteins are analyzed also in other projects of SMP-Cell, and by collaboration partners within and beyond the NGFN. The comparison and integration of results from different cellular assays being carried out in different laboratories necessitates a comprehensive description of the experimental conditions that is provided in a commonly accepted format, as well as standardized output formats for variable types of experimental data. We have initiated the establishment of such QM&S measures within SMP-Cell, as a close collaboration with other NGFN partners and with international initiatives. In view of the need for streamlined and coordinated QM&S activities, it is planned to organize a workshop with international participation that addresses these issues.
Outlook
QM&S will be of increasing importance within SMP-Cell and the NGFN, and for genomic research in general. The cost efficient generation and sharing of standardized, high-quality material, and the integration of data from different functional genomic projects necessitate comparability of experimental conditions and data formats. Standardization of assay descriptions and data formats will be the focus of our work in the foreseeable future. SOPs as well as the emerging standards for genomics experiments will continue to be published In the web-pages of the QM&S project of SMP-Cell.
Lit.: 1. Wiemann S et al. Toward a Catalog of Human Genes and Proteins: Sequencing and Analysis of 500 Novel Complete Protein Coding Human cDNAs. Genome Res. 2001 March;11(3):422-35. 2. Wiemann S et al. cDNAs in functional genomics and proteomics: The German cDNA Consortium. CRBiologies. 2003 326:1003-9. 3. Simpson JC et al. Systematic subcellular localization of novel proteins identified by large scale cDNA sequencing. EMBO Rep. 2000 1(3):287-92. 4. Wiemann S et al. From ORFeome to biology: a functional genomics pipeline. Genome Res. 2004 14(10b):2136-44. 5. Frey B et al. Demonstration of the Expand PCR System's Greater Fidelity and Higher Yield with a lacI-based PCR Fidelity Assay. Biochemica. 1995 2:8-9. 6. Arlt DH et al. Functional profiling: from microarrays via cell-based assays to novel tumor relevant modulators of the cell cycle. Cancer Res. 2005 65(17):in press.
Please visit the project's website


