
Introduction
The processes of tissue development and homeostasis require a strictly regulated balance between cell proliferation and apoptosis. Somatic cells proliferate via a determined progression through the cell cycle, which is controlled at several check points.
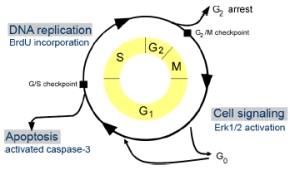 | |
| Fig 1: HTS-Assays cover cell cycle progression and apoptosis. | |
Apoptosis occurs in a wide range of physiological processes, where damaged and unnecessary cells are removed. Cancer development and tumor progression are closely associated with a dys-regulation of cell cycle control and restricted apoptosis.
Several proteins with well established function in signaling pathways take part in these cellular events. The Human Genome Project has revealed numerous further proteins of yet unknown function. We apply systematic approaches to discern information on function and potential cancer relevance to these proteins. In our approach, expression profiling data from SMP-RNA (gene expression in pathological tissue is compared with that in normal tissue) is combined with high-throughput cell-based functional assays in order to select novel targets for drug discovery.
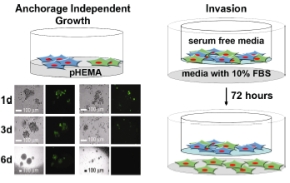 | |
| Fig 2: Assays investigating anchorage independent growth (left) and invasion (right). | |
Project Status
Assays
We have established a range of HTS cell-based assays (1) that investigate the activity of proteins during different points of the cell cycle (Fig. 1). The assays are based on effects that are induced by protein overexpression which are monitored after transfection and expression in NIH3T3 or HEK 293 cells. A proliferation assay measures the effect of an overexpressed protein on incorporation of BrdU during DNA synthesis (S) phase. The MAP kinase assay detects changes in phosphorylation of p42/p44 (ERK1/2) after transfection, in the first growth phase (G1). An apoptosis assay identifies proteins that affect the activation of caspase-underneath of that matrix. Invading cells are stimulated to degrade the matrix with their specific repertoire of enzymes and pass through that matrix and reach the chemoattractant. These cells are then harvested and counted by FACS.
Proof of principle study
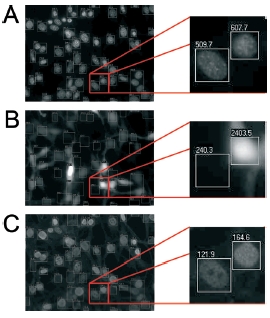 | |
| Fig 3: Comparison of BrdU incorporation between transfected an non-transfected cells. A: DAPI staining. B: Expression of the protein of interest fused to YFP. C: BrdU incorporation quantified with Cy5-labeled antibody. Fuorescence intensities are recorded for every cell individually. | |
We have implemented bioinformatics tools and databases that are suitable to analyze, store and integrate the different types of data from high-throughput experiments and to include further annotation that is based on external information. With this approach we identified 16 novel proteins that affect cell proliferation, 7 activators and 9 repressors. Out of these candidates 3 proteins influence MAPK signaling and additional 3 proteins represses anchorage independent growth.
This novel strategy combines genomics with cell biology and demonstrates its efficiency by verifying novel cancer relevant modulators of the cell cycle. For example, the gene for cDNA DKFZp434I0515 was found down regulated in kidney tumors by tumor profiling (SMP-RNA), while the gene is expressed in normal kidney tissue. Neither any functional information nor data about its effect on cell cycle regulation were available. Our assay-data showed that the protein inhibits proliferation as well as MAPK signaling, making this protein a prime candidate for further in-depth studies to investigate its role in cancer development and progression.
Current screening and throughput
Over 200 ORFs have thus far been screened for effects on proliferation, MAPK signaling, and apoptosis through flow cytometry, identifying additional cell cycle modulators. Currently some 900 ORFs are in our screening pipeline, selected through integration of protein networks and expression profiling data. Identified candidates are further validated through their effects on anchorage independent growth and invasion.
The same set of proteins is currently analyzed in an RNAi screen, where potential inhibiting effect of proteins on apoptosis induction with staurosporine is investigated (Fig. 4). That way we complement the data set resulting from the screen with overexpressed proteins.
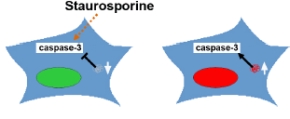 |
|
Fig 4: RNAi assay principle (left) for a screen of apoptosis inducing proteins complementary to the overexpression approach.
Outlook
The identification of novel molecular targets for drug discovery is one of the main goals of human genome projects. The implementation of high throughput technologies has enabled to collect data about novel genes and encoded proteins at a large scale. Our systematic approach combines novel high throughput cell based assay technologies with previously known data to prioritize genes and proteins that have the highest potential to be disease relevant.
Further screening of non-characterized proteins will provide a very efficient means for the identification of cell cycle relevant proteins, here emphasis will be put on those that are potential cancer drug targets. These will be validated in vitro and in vivo to select proteins of clinical relevance for further validation.
Lit.: 1.Wiemann S et al. From ORFeome to biology: a functional genomics pipeline. Genome Res. 2004 14(10b):2136-44. 2. Sültmann H et al. Gene expression in kidney cancer is associated with novel tumor subtypes, cytogenetic abnormalities and metastasis formation. Clin Canc Res. 2005 Jan 15;11(2 Pt 1):646-55. 3. Arlt DH et al. Functional profiling: from microarrays via cell-based assays to novel tumor relevant modulators of the cell cycle. Cancer Res. 2005 65(17):in press. 4. Wiemann S et al. Toward a Catalog of Human Genes and Proteins: Sequencing and Analysis of 500 Novel Complete Protein Coding Human cDNAs. Genome Res. 2001 March;11(3):422-35. 5. Simpson JC et al. Systematic subcellular localization of novel proteins identified by large scale cDNA sequencing. EMBO Rep. 2000 1(3):287-92.


