
Introduction
Determination of the sub-cellular localisation and dynamics of any protein is an important first step towards the understanding of its function in a cellular context. Proteins of functional networks must co-localise in order to interact with each other and changes in protein localisation in response to external stimuli reflect on changes of activities.
We have developed and applied a systematic pipeline to determine the sub-cellular localisation of the novel proteins identified by the German cDNA Consortium, and of proteins relevant to collaborators within the NGFN1. The proteins under investigation are systematically analysed by the bioinformatics tools that we have previously developed for this purpose (1,2). After such analyses all the novel cDNAs are tagged at their C- or N-terminal end with spectral variants of the green fluorescent protein (GFP). Finally, the tagged cDNAs are expressed in living cells and the sub-cellular distribution of the fusion proteins is documented by fluorescence microscopy.
For those proteins where the distribution is difficult to associate clearly with sub-cellular structures or organelles, co-localisation experiments with established markers are performed. Integrating the bioinformatics information with the experimental results obtained for N- and C-terminal fusions is then used to determine the localisation of the respective proteins. The localisations obtained by this protocol could be confirmed in about 85% by immuno-cytochemistry using antibodies raised against a fraction of the proteins under investigation. The results and bioinformatics information on the proteins localised so far is publicly available in the LIFEdb database (http://www.dkfz.de/LIFEdb).
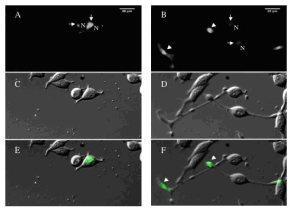 | |
| Fig 1: Images of the first and last time-point of a time-lapse sequence investigating protein dynamics in PC12 cells. PC12 cells were transfected with a GFP-tagged novel cDNA for 16h and subsequently imaged at 20min time-intervals for further 24h after NGF stimulation. A, C, E show the first time-point of the sequence immediately after NGF addition; B, D, F show the time-point 24h later. Arrows in A and B point to Golgi localization, arrowheads in B and F point to the end of neuritis that have grown in response to NGF stimulation. A strong accumulation of the tagged protein in the end of neurites upon NGF stimulation is observed. | |
Project Status
The goal of this sub-project within SMP CELL is to GFP-tag 1,200 new cDNAs, to localise the encoded proteins in living cells and also to study the dynamics of the localisation of all the so far cloned proteins (936 clones generated in the framework of NGFN1 and the new 1,200 cDNAs tagged in this project) in response to mitogenic stimuli and during cell differentiation. To this end we have developed during the course of this project software packages that allow us the determination of the dynamics of fluorescent structures by advanced tracking methods such as vector flow analysis.
We have further adapted the reverse transfection of cells on cDNA arrays (3) to obtain the necessary throughput and reproducibility in localisation analyses. We have also adapted our previously developed high content screening microscopy technology (4) for automated data acquisition and analysis of cells on cDNA arrays. We have started to use PC12 cells to investigate the changes in localisations of our GFP-tagged proteins during neurite outgrowth by time-lapse microscopy. To this end we have established a strain of this cell type that can be stimulated to grow long neurites already between 12 and 24h after addition of Nerve Growth Factor (NGF) (see Fig.1). Therefore, this strain is convenient to monitor neurite outgrowth as a marker for neuronal differentiation by time-lapse microscopy.
Since the start of the project in November 2004 we could tag and localise 300 more novel proteins and have analysed their dynamics of localisation in PC 12 cells after NGF stimulation. We will also determine protein dynamics in unperturbed HeLa and Vero cells. We will also monitor protein dynamics after depletion of cellular cholesterol in order to obtain possible candidates involved in the regulation of cellular cholesterol levels. Together with the group of Stefan Wiemann at the DKFZ in Heidelberg we will make available all the data on the dynamics of protein localization in the LIFEdb database. Most importantly, the steady state localization as well as the protein dynamics data will serve as first selection criteria for functional studies addressing cholesterol-dependent membrane transport and neuronal differentiation.
Lit. 1. Bannasch D et al. LIFEdb: a database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system. Nucleic Acids Res. 2004 32:D505-8. 2. Liebel U et al. "Harvester" a fast meta search engine of human protein resources. Bioinformatics 2004 12:1962-63. 3. Erfle H et al. siRNA cell arrays for high content screening microscopy. Biotechniques 2004 37:454-458, 460, 462. 4. Liebel U et al. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 2004 Nov; 554(3): 394-8
Please visit the project's website


