
Introduction
Protein variants as a result of alternative splicing have been shown to be associated with human diseases, such as cancer (1) and several neurodegenerative disorders (2). Therefore the correlation of splice form variation and its protein function with relevance to human diseases is of great importance.
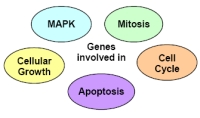 | |
| Fig 1: Focus on genes that are involved in these cancer-relevant pathways. | |
Second, the splice variant exists in the coding region and shows at least one exon difference (Fig.2).
Our aim is the amplification of the complete open reading frame (ORF) of splice variants of these genes and their cloning into a shuttle vector system (Gateway®). This allows us to express the ORFs of the splice variants N- and C-terminally fused to the yellow fluorescent protein (YFP). The YFP-tagged splice isoforms will then be analysed by the high throughput functional assay (3) within SMP-Cell.
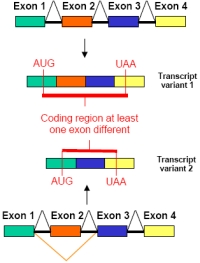 | |
| Fig 2: Transcript variants that show at least one exon difference and thus encode variant proteins. | |
Project Status
Search for genes with splice isoforms
We started with all Unigene clusters and selected those clusters which contained more than one validated mRNA (e.g. Refseq). This means that there exists at least two splice variants for the respective gene. Moreover, we considered only those validated mRNAs which were represented by a full length clone. From these selected genes we mapped the encoded proteins on pathways and selected specifically such that were identified to be involved in those pathways described in Fig. 1. In the next selection step we focused only on those genes of which different functions of the splice variants have not been published yet.
Validation of splice variants
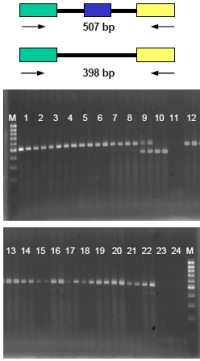 | |
| Fig 3: Example for the validation of splice variants in different human tissues by RT-PCR. Top:Transcript variant 1; below: Transcript variant 2. M = ladder 1 = thyroid gland 2 = thymus 3 = thymus 4 = tonsil 5 = tonsil 6 = lymph knots-T 7 = marrow 8 = spinal cord 9 = brain 10 = brain 11 = H2O 12=heart 13 = heart 14 = lung 15 = lung 16 = lung-tumor 17 = Mamma 18 = Mamma 19 = breast-tumor 20 = muscle 21 = muscle 22 = retina 23 = H2O 24 = H2O | |
Outlook
This subproject describes for the first time the cloning of complete ORFs that carry the alternative splice isoforms and therefore provides access to the transcriptional isoforms for downstream functional analysis. 507 bp Transcript variant 2 Our final goal is to identify splice variants that show different effects in functional assays.
These variants will then be further characterised. One interesting question will be whether these splice variants bind to different interaction partners. This could be analysed by yeast-two-hybrid-interactions and co-immunoprecipitation. Furthermore, the expression profile of these splice variants in different tissues (also control tissue versus disease tissue) might be of central interest.
Lit.: 1. Roy M et al. Evidence that public database records for many cancer-associated genes reflect a splice form found in tumors and lack normal splice forms. Nucleic Acis Res. 2005 Sep 7;33(16):5026-33. 2. Lee CJ and Irizarry K. Alternative splicing in the nervous system: an emerging source of diversity and regulation. Biol Psychiatry. 2003 Oct 15;54(8):771-6. 3. Arlt D et al. Functional Profiling: From microarrays via cell-based assays to novel tumor relevant modulators of the cell cycle. Cancer Res. 2005 Sep 1;65(17):7733-42 4. Gupta S et al. Strenghts and weakness of EST-based prediction of tissue-specific alternative splicing. BMC Genomics. 2004 Sep 28;5(1):72 5. Gupta S. et al. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics. 2004 Nov 1;20(16):2579-85
Please visit the Project's Website


