
Introduction
Understanding the pathogenesis of human disease is an essential prerequisite for developing efficient pharmaceutical compounds and therapies. Many human diseases arise by distortions in signalling mechanisms of complex nature. Signal cascades are no isolated entities, but interconnected. Moreover, there is only a fairly small number of signal cascades which are utilized over and over again in a rather large number of processes in the adult organism as well as in embryonic development. And they are modulated by many modifiers resulting in various cellular responses. To date it is not clear, except for a few known cases, how signal cascades are interconnected and what distortions exactly lead to disease. For that reason it is important to study signalling mechanisms, including cellular readouts, as a whole, and begin to elucidate the genetic networks involved in biological processes and in disease. With respect to cellular readouts, more specifically the cellular response in terms of target gene activation downstream of signalling events, which is essential information for deriving regulatory networks, genetic and genomic research is still in its infancy. So far, regulatory networks have only been described on a small scale except for yeast (for review see Wyrick and Young, 2002; Lee et al. 2002). However, time is ripe for tackling this problem on a larger scale also in higher vertebrates, though a genome-wide approach has to be limited to elucidating regulatory networks controlling particular well defined processes.
The mid-gestation mouse embryo, which is subject of multiple signalling processes, offers an in vivo system to study signalling mechanisms and cellular readouts in the natural context. We have decided to focus on signalling mechanisms downstream of Wnt/#-cat, FGF and TGF#1/BMP, which are strongly related to cancer and metastasis formation, to investigate regulatory networks controlling epithelial-mesenchymal transition (EMT) in the midgestation mouse embryo. EMT is an essential step in metastasis formation and in organogenesis (Savagner 2001). By elucidating the regulatory network controlling EMT in the embryo (in vivo) we are confident to derive information essential to understand metastasis formation, which cannot be analysed in vivo (in the patient). This may eventually lead to the identification of drug targets and pharmaceutical compounds suitable for preventing metastasis formation in humans.
One essential component in building regulatory networks is the identification of target genes activated by signalling events. Bioinformatic means are suitable to predict promoter regions (reviewed by Ohler and Niemann, 2001) and to identify potential targets (immediate early response genes) by searching for putative binding sites for transcription factors activated by signal cascades, in the putative promoter regions. Target genes downstream of signal cascades can also be identified by gene expression analysis of mutant embryos in which the signal cascade has been disrupted. A third set of data can come from high resolution expression analysis, identifying genes co-expressed with the signal molecules controlling the cascades under study. The combination of all three methods will provide a degree of accuracy in predicting real target genes which can only be surpassed by experimental prove.
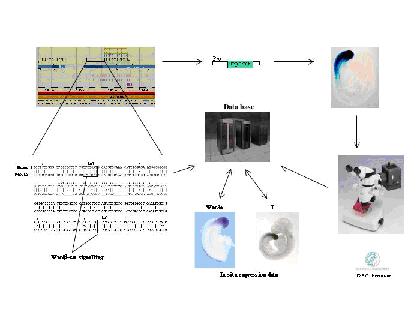 |
| Fig 1:Promoter analysis in silico and in vivo. Predicted control factor-target realtionships are assayed in vvio and correlated by co-expression of the respective genes in the embryo |
In this project we will combine data derived from bioinformatic promoter analysis (SMP DNA/Vingron), high-resolution gene expression analysis in mouse embryos (SMP RNA/Herrmann), and knock-down phenotype analysis (SMP RNAi/Herrmann) to predict regulatory networks controlling EMT downstream of Wnt/#-cat, FGF and TGF#1/BMP.
The goal of this subproject is to experimentally verify the promoter predictions derived from bioinformatic analyses, in the embryo using eGFP as reporter. The activity of the promoter fragment will be compared to the expression pattern of the gene under study to reveal potential differences, since the promoter fragment may only reflect a subset of the expression sites of the gene. The outcome is correlated with the predicted transcription factor binding sites on the promoter fragment used, and with the expression patterns of the transcription factors predicted to bind to the promoter (SMP_RNA/Herrmann).
We will introduce a number of predicted promoter fragments driving eGFP expression into ES cells and determine the expression pattern of the reporter in tetraploid embryo/ES cell chimaera at various stages of embryonic development. Tetraploid embryos used as recipients for ES cell clones derived in vitro allow the formation of chimaeric embryos whose embryo proper is exclusively derived from the ES cells, while the host cells are restricted to extra-embryonic tissues (Nagy et al. 1993). Thus tetraploid embryo/ES cell chimaera allow the analysis of reporter constructs introduced into ES cells in basically hemizygous embryo propers.
Promoter fragments selected for analysis will be derived from genes downstream of Wnt/#-cat, FGF and TGF#1/BMP signalling. The experimental outcome will be correlated with high resolution gene expression data obtained from transcription factors predicted to control the gene under study, and with expression data obtained from RNAi experiments in which activators of the promoter under study have been knocked down. This will allow identification of regulator-target interactions and building of regulatory networks.
Outlook
Understanding regulatory networks controlling signalling mechanisms involved in embryonic processes as well as in the pathogenesis of human disease is a challenge to state of the art systems biology approaches requiring tight collaboration of a number of groups obtaining various data sets. In vivo promoter analysis provides a data set necessary for verifying in vivo essential pieces of a puzzle constituting a regulatory network. Thus, in vivo promoter analysis is an essential component in the concerted efforts of deciphering regulatory networks.
Lit.: 1. Lee TI, Young RA et al. (2002). Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799-804. 2. Nagy, A., Rossant, J., et al. (1993). Derivation of completely cell culture-derived mice from early passage embryonic stem cells. Proc.Natl.Acad.Sci. USA 90, 8424-8428. 3. Ohler U, Niemann H. (2001) Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet 17, 56-60. 4. Savagner, P. (2001). Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. BioEssays 23, 912-923. 5. Wyrick J.J. and Young R.A. (2002) Deciphering gene expression regulatory networks. Curr Opin Genet Dev. 12, 130-6.


