
Introduction
Haplotypes present an important tool in genetic epidemiological association studies. They summarize the information of several SNPs and additionally incorporate phase information. It is important to know the phase of haplotypes because it provides information about the passing on of genes over the generations and is widely believed to play a role in association analyses. However, it is technically elaborate to determine haplotypes molecularly. Therefore, haplotypes are mostly reconstructed with statistical methods. The reduced work effort is paid for with an additional source of error in the reconstruction process.
Many problems must be dealt with in haplotype-based studies. Besides the reconstruction error in general, the choice of SNPs that form the haplotypes is essential. Haplotype length and SNP density must be considered. It is clear that densely spaced SNPs are desirable for thorough haplotype analysis, but longer haplotypes also mean more work in the laboratory. Moreover, high linkage disequilibrium (LD) leads to redundancies so that some SNPs may contribute little or no information. Haplotype tagging methods try to account for this tradeoff between information content and laboratory work by choosing a subset of the SNPs that “tag” the haplotype, i.e. that contain most of the information without being too long.
In this project, we consider properties of haplotypes in association studies. We evaluate the effect of SNP density on haplotype analyses. HapMap-SNPs serve as reference against high-resolution SNPs. To evaluate the role of haplotype reconstruction, we compare several statistical methods against each other and against molecularly obtained haplotypes. We also concentrate on the differences between microsatellite and SNP markers and how they complement each other.
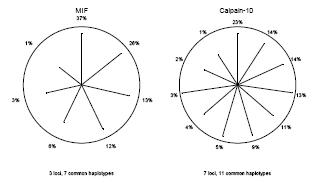
Fig 1: These star plots for the genes MIF and Calpain-10 show the error measure R2. Each radius stands for one haplotype, and the length of the radii indicate the R2 of the gene. The haplotypes are sorted clockwise by their frequency, beginning at the top with the most frequent one. Haplotypes with long radii have a high R2 value (i.e. are estimated fairly well). Haplotypes with frequencies less than 1% are omitted.
The topic of haplotypes in association studies is highly diverse. In our recent studies, we mainly assessed the importance of statistical analyses concerning the nature of the reconstruction error and their characterization via different error measures on the one hand, and SNP density and its effects on LD structures, block partitioning, tagging SNPs and haplotype reconstruction on the other.
Error Measures in Haplotype Reconstruction
Statistical reconstruction of haplotypes is necessarily subject to errors. We evaluated a broad range of error measures that complement each other for a complete description of reconstruction performance. Theoretically derived models are useful for the observation of general properties of the error. But we also used scenarios based on real genotype data to observe the range of the error in realistic settings. The data was obtained from 704 individuals from the KORA (Cooperative Health Research in the Region of Augsburg) S4 survey.
Our analysis comprised the comparison of haplotype reconstruction methods based either on the expectation-maximization (EM) algorithm or on Bayesian methods. We evaluated the error based on simulations and on analytical derivations.
We performed a systematic investigation of a set of measures that cover different aspects of insecurity in haplotype reconstruction. Error measures can be classified into such that use the most likely haplotype pair (“best guess” haplotypes) and into such that use the estimated number of haplotypes for each individual. Both methods have advantages and disadvantages and it must be carefully considered which method to use in an association study. A second way to characterize measures is whether they give one combined value for the haplotype reconstruction, or if single values are given for each single haplotype. Which approach is better suited in a particular situation depends on the aims of a study. In association studies it may e.g. be more interesting to get one value per haplotype, because single haplotypes are viewed as potential risk factors. In a study comparing the performance of two haplotype reconstruction methods, however, an overall measure for all haplotypes may be the better choice. Figure 1 shows the error measure R2, which evaluates the haplotypes using the expected number of copies, for the genes MIF and Calpain-10.
The influence of the reconstruction error on association studies is another topic we treat in our analyses. We also perform simulations on the significance of genotyping errors for the haplotype reconstruction process. Further, we treat the efficieny of tagging SNPs on the example of the adiponectin encoding APMA gene.
LD Patterns in European Populations
The LD pattern is an essential factor in genetic association studies, where disease-causing variants are identified by neighbouring markers in high LD. We compared the LD patterns in eight distinct European populations selected along a line from north to south. Altogether we analyzed four genomic regions containing candidate genes for complex traits. In general, we observed a conservation of LD patterns across European samples. Nevertheless, shifts in the positions of the boundaries of high-LD regions can be demonstrated between populations, when assessed by a novel procedure based on bootstrapping. Transferability of LD information among populations was also tested. In two of the analyzed gene regions, sets of tagging SNPs selected from the HapMap CEPH trios performed surprisingly well in all local European samples. However, significant variation in the other two gene regions predicts a restricted applicability of CEPH-derived tagging markers. Simulations based on or data set show the extent to which further gain in tagSNP efficiency and transferability can be achieved by increased SNP density.
Outlook
The evaluation of the haplotype reconstruction error with different error measures and the effects on association analyses will be continued. Chosen genes of the KORA population will be further analyzed in light of their LD structures. The determination of haplotype tagging SNPs and the partitioning into high-correlation blocks are of special interest.
The effects of SNP density on LD structures are further pursued. Along with changes in LD structures, the choice of tagging SNPs and the boundaries of haplotype blocks can change. The reconstruction of haplotypes is also dependent on SNP density and is also subject of our analysis.
The performance of various haplotype reconstruction methods has been evaluated by means of simulations based on KORA haplotype frequencies. These methods will also be compared to molecular haplotypes. The previous observations about properties of a variety of error measures will be the basis for the new analysis. Besides the quantification of the error, the effects on tagging SNP selection are observed. Different algorithms for block partitioning will be applied in this framework.
A further topic is the analysis of microsatellite markers and SNP markers and the question how they relate to each other. We examine the different aspects in association studies that are addressed by the two approaches.
Lit.: 1. Müller JC, Lohmussaar E, Mägi R, Remm M, Bettecken T, Lichtner P, Biskup S, Illig T, Pfeufer A, Luedemann J, Schreiber S, Pramstaller P, Pichler I, Romeo G, Gaddi A, Testa A, Wichmann HE, Metspalu A, Meitinger T. Linkage Disequilibrium Patterns and tagSNP Transferability among European Populations. Am J Hum Genet. 2005 Jan 6;76:387-98. 2. Heid IM, Lamina C, Bongardt F, Fischer G, Klopp N, Huth C, Küchenhoff H, Kronenberg F, Wichmann HE, Illig T. How About the Uncertainty in the Haplotypes in the Population-Based KORA Studies. Gesundheitswesen. 2005;67(S1):132-6. 3. Illig T, Bongardt F, Schöpfer-Wendels A, Huth C, Heid I, Rathmann W, Martin S, Vollmert C, Holle R, Thorand B, Wichmann HE, Koenig W, Kolb H, Herder C; KORA Study Group. Genetics of Type 2 Diabetes: Impact of Interleukin-6 Gene Variants. Gesundheitswesen. 2005;67(S1):122-6. 4. Illig T, Bongardt F, Schöpfer A, Müller-Scholze S, Rathmann W, Koenig W, Thorand B, Vollmert C, Holle R, Kolb H, Herder C; Kooperative Gesundheitsforschung im Raum Augsburg/Cooperative Research in the Region of Augsburg. Significant Association of the Interleukin-6 Gene Polymorphisms C-174G and A-598G with Type 2 Diabetes. J Clin Endocrin Metab. 2004 Oct;89(10):5053-8. 5. Heid IM, Vollmert C, Hinney A, Döring A, Geller F, Löwel H, Wichmann HE, Illig T, Hebebrand J, Kronenberg F; KORA Group. Association of the 103I MC4R Allele with Decreased Body Mass in 7937 Participants of two Population Based Surveys. J Med Genet. 2005 Apr; 42(4):e21. 6. Weidinger S, Klopp N, Wagenpfeil S, Rummler L, Schedel M, Kabesch M, Schafer T, Darsow U, Jakob T, Behrendt H, Wichmann HE, Ring J, Illig T. Association of a STAT 6 Haplotype with Elevated Serum IgE Levels in a Population Based Cohort of White Adults. J Med Genet. 2004 Sep; 41(9):658-63. 7. Justenhoven C, Hamann U, Pesch B, Harth V, Rabstein S, Baisch C, Vollmert C, Illig T, Ko YD, Bruning T, Brauch H. ERCC2 Genotypes and a Corresponding Haplotype are Linked with Breast Cancer Risk in a German Population. Cancer Epidemiol Biomarkers Prev. 2004 Dec; 13(12):2059-64.


