
Introduction
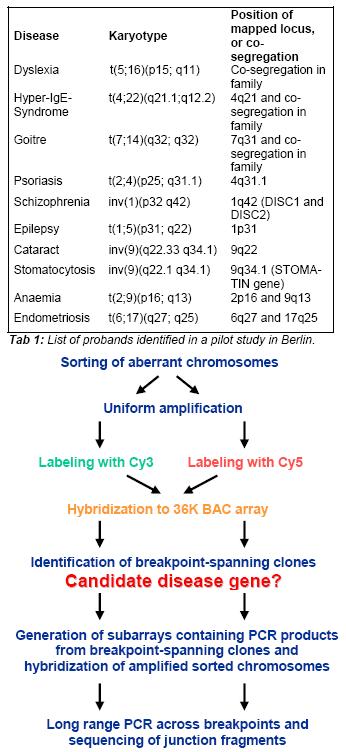
Fig 1: Flow-chart illustrating our novel protocol for large scale breakpoint mapping and gene identification.
Disease-associated balanced chromosomal rearrangements (DBCRs) such as translocations or inversions are valuable resources for the identification of disease-causing genes by molecular cytogenetic breakpoint analysis. The systematic study of DBCRs has led to the elucidation of numerous disease genes, and in recent years, we have identified more than 30 disrupted or otherwise inactivated genes in about 150 patients with DBCRs (1-5, unpublished results). In view of these encouraging results, we extended our systematic study of DBCRs to rearrangements that are associated with late-onset or complex diseases. De novo balanced chromosome rearrangements associated with inborn disorders or diseases manifesting during the first years of life are rare (approximately 1/25,000 newborns). In contrast, familial translocations or inversions are much more prevalent (approximately 1/1,000 newborns). It can be assumed that many of these usually healthy carriers of balanced chromosome rearrangements (BCRs) also carry disrupted genes since about 30% of the human chromosomes are covered by genes (including introns), and chromosome breakpoints seem to be evenly distributed. The majority of these disrupted or otherwise inactivated genes are apparently recessive and do not cause any early-onset disorders. However, little attention has been paid so far to the possibility that the disruption of such genes might predispose to complex, late-onset disorders such as neurodegenerative, cardiovascular or metabolic diseases, cancer and infertility. In collaboration with other cytogenetic laboratories and institutes for Human Genetics, we have performed a questionnaire-based survey among adult BCR carriers to identify suitable probands for breakpoint analysis . Extrapolation from an analogous Danish study (N. Tommerup and I. Bache, unpublished results) indicates that in Germany roughly 20,000 individuals should be known carriers of BCRs, and many of these should be identifiable by systematic screening of cytogenetic archives.
Project Status
Associations between BCRs and disorders reported in the questionnaire are considered as probably real if the disorder co-segregates with the BCR in a family; if one breakpoint is located at a previously identified locus for the respective disorder; or if several unrelated patients with the same disorder have a breakpoint in the same chromosome region. A pilot study in the Berlin area revealed several probands fulfilling these criteria (Tab. 1). One example is a patient with psoriasis and a breakpoint in 4q31 within a major locus for psoriasis as determined by a recent meta-analysis of linkage data. In this case, a plausible functional candidate gene for psoriasis has been shown to be affected by the translocation. We also investigate patients with amyotrophic lateral sclerosis carrying constitutional balanced chromosome rearrangements (6;7). Likewise, chromosomal breakpoints of two individuals with dyslexia, both carrying familial balanced chromosomal rearrangements that co-segregate with the disorder, are currently under study. Chromosomal breakpoints of a familial balanced translocation that co-segregates with malignancies of the lymphatic system involving chromosomes 1 and 13 are also being examined.
Fig 3: Flow-sorted DNA from chromosomes 1 and 13 was hybridised to a high resolution subarray CGH with multiple 800 bp fragments for fine mapping of the translocation breakpoint. With this novel protocol breakpoints are mapped to a region of about 4-6 kb in a single hybridisation experiment.
We have previously developed a 36,000 BAC CGH array, which has been successfully used for investigations in more than 400 patients with suspected or proven unbalanced chromosome rearrangements, and our laboratory is one of the few laboratories worldwide where this technique is fully operational. This 36K chip enables us to detect sub-microscopic deletions or duplications with a resolution of up to 100 kb. The time-consuming process of identifying breakpoint-spanning BAC clones by FISH and the subsequent mapping of breakpoints using molecular techniques has been a major limitation in the large scale study of DBCRs and BCRs. To overcome this obstacle, we have established a novel protocol allowing rapid fine-mapping and sequencing of the breakpoint regions in a large number of cases (see Fig.1). This novel protocol combines flow-sorting of aberrant chromosomes and high-resolution array CGH. In each case, both breakpoints are mapped to about 100-150 Kb in a single hybridization experiment. As an example, Fig 2 shows the results of array CGH hybridisation using sorted chromosomes from of a familial translocation between chromosomes 2 and 12.
Subarray CGH and long-range PCR
In order to map the breakpoints more precisely, subarrays carrying multiple, evenly spaced PCR-amplified DNA fragments from the breakpoint-spanning BAC clone are used. Figure 3 shows the results of a subarray CGH for a breakpoint-spanning clone from chromosome 13. Finally, long range PCR and sequencing are employed to characterize the breakpoint region at the nucleotide level.
Fig 2: Array CGH result for aberrant chromosomes 2 and 12 displayed by CGHPro (7). For detection of translocation breakpoints, flow-sorted DNA from chromosomes 2 and 12 was hybridised to a 36K BAC clone array. The ratios of the clones are plotted in a size dependent manner along the chromosome ideograms. The green and red lines represent log2 ratios of 0.3 (gains) and –0.3 (losses), respectively.
This new technology has removed the major obstacle for large-scale analysis of DBCRs and BCRs and allows rapid identification of candidate disease genes. Array CGH based breakpoint mapping is available to other groups on a collaborative basis. At least theoretically, by including all known probands with BCRs in Europe, this effort could lead to a dense functional map of the entire human genome.
Lit.: 1. Kalscheuer et al. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Gen 2003 Jun; 72(6): 1401-11. 2. Shoichet et al. Mutations in the ZNF41 gene are associated with cognitive deficits: identification of a new candidate for X-linked mental retardation. Am J Hum Genet 2003 Dec 73(6): 1341-54. 3. Dlugaszewska et al. Breakpoints around the HOXD cluster result in various limb malformations. J Med Genet. 2005 Jun 24; [Epub ahead of print]. 4. Borg et al. Disruption of Netrin G1 by a balanced chromosome translocation in a girl with Rett syndrome. Eur J Hum Genet. 2005 Aug;13(8): 921-7. 5. Shoichet et al. Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations, and microcephaly. Hum Genet. 2005 Aug17; [Epub ahead of print]. 7. Meyer et al. High rate of constitutional chromosomal rearrangements in apparently sporadic ALS. Neurology. 2003 Apr 22;60(8): 1348-50. 6. Prudlo et al. Chromosomal translocation t(18;21)(q23;q22.1) indicates novel susceptibility loci for frontotemporal dementia with ALS. Ann Neurol. 2004 Jan;55(1): 134-8. 7. Chen et al. CGHPRO -- a comprehensive data analysis tool for array CGH. BMC Bioinformatics. 2005 Apr 5;6(1): 85.


