Introduction
The basic idea of the RZPD is the use of common standardised material of proven high quality, in combination with a database to take up the most different types of data generated with the materials distributed. RZPD has been very successful over the last 10 years with the products developed, imported and distributed, being the world’s largest service and distribution centre for genome research.
Due to the genome information that is increasingly becoming available, RZPD is currently redefining its view on resources to integrate various external platforms, tools and databases. RZPD operates various databases using different relational database management systems. The databases serve different purposes, e.g. representing scientific, logistic or accounting data or a combination of these data types.
Focus of Research & Development
A flexible system for data integration across a wide variety of data and organisms, the GenomeMatrix, has been developed in collaboration with the Max Planck Institute for Molecular Genetics (Hans Lehrach, Martin Vingron, www.genome-matrix.org). To extend this concept, and to provide further sophistication in data integration across the platforms, a consolidation of the different databases and computational tools has been accomplished.
A gene-based relational database (MASI) including freely accessible database at RZPD that are linked to international databases was implemented. Due to the current lack of standards within the NGFN for data acquisition, formats, analysis, integration, and display, a tool to query independent and physically separated databases containing heterogeneous data formats is required. The GenomeCubel will extend the view on the genome to a more complex view, in particular biomedical aspects -complemented by the development of new tools and materials.
Project Status
The necessity to find optimal clones for various research purposes led to RZPD's “GenomeCube” concept by focusing on its ability to link information about genes to biological material. For each gene of a species RZPD is able to provide multiple clones and experimental information of a different functional type.
Thus, the GenomeCube differs from other international molecular biology databases with respect to the ability to integrate and store various data types. Data types that can be stored in the database include sequences (incl. annotations, trace data), Blast search results, in situ hybridization images, hybridization data from array screenings including description of probe, radiation hybrid mapping data, chromosomal positions of clones, gene cluster of various organisms and cluster algorithms, gene names of Unigene cluster, gene expression data of micro- or macroarrays, cloning parameter, hybridization protocols, complete nonredundant gene sets, siRNA probes, and alternative clone naming and conversion.
1) Data integration using GenomeCube, GenomeMatrix, and MASI
The GenomeCube™ combines biological information from several sources, integrates them locally and provides links to and from other databases.
So far, GenomeCube™ integrates 281.567 genes (based on NCBI’ UniGene Cluster) and 4.200.000 clones and clone-derived products for human, mouse, rat, Xenopus, zebrafish and Arabidopsis.
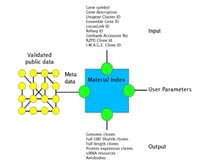
The easy-to-use database architecture of GenomeMatrix is aimed to allow the integration of diverse types of data, e.g. data like gene description according GeneOntology, related diseases, protein structures, protein-protein interactions, metabolic pathway information, in situ-hybridisations, knock-out mutants and biological material for a gene (e.g. Unigene clones, complete coding sequence clones, etc).

The MASI database (InSilico, Wien) is a compilation of public biological databases into a locally available relational database management system (RDBMS). The increasing spectrum of information provided, covers all relevant and interlinked sources of genetic, proteomic and metabolomic data. Therefore, particular biological entities cited at different sources become comparable and thus provide more comprehensive information.
The aim of the integration project of GenomeCube, GenomeMatrix and MASI is to provide middleware technology to allow users to discover biomedical knowledge that are based on widely distributed data. In effect, the project aims to provide a bridge between the scientific community performing analysis using biological materials and the community who generates the data.
2) Gene expression data analysis and ExpressCube
ExpressCube is a public repository for microarray data located at the RZPD. It stores data generated in gene expression experiments in accordance with the recommendations of the Microarray Gene Expression Data (MGED) Society. Experiments can be queried by certain criteria and are displayed in a user-friendly way using the MAGE terminology. Data can be downloaded in matrix format and can be further examined using RZPD's automated analysis pipeline, which combines statistical methods and functional annotations for the most promising genes. As part of a European project, data submitted in a MIAME compliant way will also be forwarded to ArrayExpress, the international microarray database located at the European Bioinformatics Institute.
The methodoly behind the ExpressCube is the development of data flow processes or pipelines that represent the transformation of data from one form to another using a variety of different services.
The standardization of the analysis of gene expression profiling experiments is the only way to make such experiments comparable to each other and to control the quality of the experiment results. For this purpose the RZPD is establishing an automated analysis pipeline.
This includes some standard statistical methods
- "M versus A analysis"
- "t-test" or "Anova" including methods for multiple testing corrections
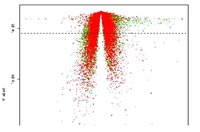
- Volcano Plot
- "Principal Component analysis"
- Clustering
as well as further annotations for the statistically most promising genes from the first step
This leads to an extended protocol for gene expression profiling experiments, which enables scientists to evaluate their experiments at a glance from a set of standardized parameters determined by RZPD's expertise.
The pipeline is designed for easy use. Data from Affymetrix experiments can be uploaded (CEL files) or gene/clone tables, that are formatted as hybridisation expression values per column. The pipeline is configured with default parameters, i.e. the analysis can be started directly after the upload. It is also connected to the ExpressCube#, i.e. experiments can be directly loaded into the pipeline for automated analysis.
The results are returned to the user as a link to a webpage and PDF Document holding the results.
The development of the pipeline is based on open source software, e.g. "R", the language for statistical computing and graphics and well developed information technology methods, e.g. "ANT" (Another Neat Tool) which is a Java-based build tool, similar to "make". ANT can be extended using Java classes and the configuration files are XML-based. Thus, it is particularly well suited for setting up an automatic pipeline.
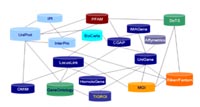
3) Data interpretation using meta-data approach and topic maps
Biomedical research process is composed of many stages: gene activity validation, exploration of signal transduction, identification of protein-protein interactions, target finding, and clinical trials. Each stage requires different knowledge and information from a wide variety of data resources, such as medical, gene, protein and compound databases. It is, however, very difficult to bridge these databases since there are large semantic gaps among their background knowledges (medical science, molecular biology and pharmaceutics).
There are two dimensions to accessing information, where the information is and how to interpret. Or, in other words, knowledge is data plus interpretation.
Topic maps provide a standard way to represent and interchange information assets, such as list of terms, ontologies, and vocabularies. The work on topic maps began in 1991, was finalized as ISO 13250 standard in 1999, and published in 2000 by World Wide Web Consortium (W3C). Using the meta-data, RZPD develops a system based on topic maps, which enables RZPD to access databases online through a knowledge representation technology: GenomeCube Service.
Outlook
The concept of GenomeCube reflects the demand to solve the big data problem of genomics by finding a balance between innovation and commonality. On one hand the scientist needs gene-based materials, on the other hand the biomedical researcher needs an approach that relies on complexes of interacting gene products involved in complex cellular behavior.
GenomeCube uses molecular data mining and knowledge discovery to correlate the scientific assurance of the data as a crucial phase in improvement of providing this integrated framework of data handling, large-scale analysis, manipulation and interpretation.


