Introduction
The availability of complete genome sequences for human and other organisms is expected to change the way we formulate and address biological questions. With nearly all genes in hand, the conventional approach of studying one gene at a time can now be complemented by more global or integrative approaches that consider all genes at once.
The complexity inherent to simultaneously considering tens (or hundreds) of thousands of proteins to formulate integrative biological questions is such that biologists will increasingly need various maps that indicate (even crude) information on protein function.
One of the many ways functional biology will become better and better over time is by improving the quality of the genomic data itself. A small proportion of the human genome still remains to be sequenced and the number of genes is still unclear. In addition, the rate of false positives and false negatives for exon predictions could be high, especially with the problem of frequent alternatively spliced variants.
Many methods for high-throughput, experimental elucidation of gene function (functional genomics) depend on the availability of full coding cDNA clone collections, so-called full ORF (open reading frame) clones. These clones provide access to the protein-coding ORFs and facilitate expression of large numbers of proteins in the native form or as fusion proteins. The value of full ORF cDNA clone collections (ORF clones) has now been amply demonstrated by studies in model organisms and human, in particular in the area of protein expression (1, 2), protein interaction mapping using methods based on yeast two-hybrid (3), mass spectrometry (4), or large scale functional screening (5).
Results/Project Status
Cloning human and mouse ORFs
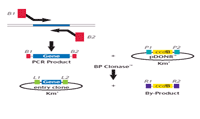
To extend the coverage of full ORF clones, we have developed a pipeline which exploits knowledge of gene structure based on genomic sequence and cDNA Sequence information as well as gene prediction.
We define potential full length clones harbouring sequence of 5’ UTR, coding sequence and 3’ UTR. These clones are defined by sequence analysis of 5’ and 3’ ESTs or the complete clone sequence where available. RZPD does make use of any clone source available, incl. IMAGE (6), German cDNA Consortium (7) and MGC (8). Whenever the informatic tools predict that a clone has potential for full coding content, we use gene-specific amplification to subclone the ORF using the Gateway recombination technology (see fig. 1).

Due to the inherent step of PCR we have introduced strict quality control steps that validate the insert size as well as the insert sequence of the final full ORF clones. Because in many applications full ORF clones are used for protein expression, it is important that the clones are annotated to
high standards and that all potential mutations are identified. Therefore we sequence verify at an error level that is better
than 1/10.000 bases, and annotate any base pair and amino acid exchanges that are coded by each individual clone (see fig. 2).
All clones that pass the QC process are submitted to EMBL Bank, and are available for distribution.
In the past we have deposited more than 3.600 ORF sequences in public databases, and will create another 3.000 within this project. We have teamed up with the Harvard Institute of Proteomics in order to speed up the overall process and reduce redundancy in cloning and sequencing. This very active collaboration might allow the RZPD to double the milestones described in the SMP, and should deliver up to 7.000 ORF clones over the next two years.
Status of the subproject “Full ORF cloning”
More than 2.100 full ORF clones have been analysed. In total 1858 clones have undergone sequence analysis. Our failure rate in cloning designated ORFs is at 6%. Sequence verification identified that in another 5% of the cases we detect frame shifts or mutations in essential amino acids. A final 3% of the clones show irregular growth after freezing and strain building. Therefore we expect to submit to EMBL Bank at least 1.600 human full ORF sequences by the end of 2005. In parallel, the clones will be completely annotated and incorporated into RZPD’s search tool GenomeCube (available at http://www.rzpd.de/products/orfclones/).
At RZPD, various applications have been implemented for the use of full ORF clones, including high parallel protein expression, comparison of in vivo and in vitro expression (E.coli and wheat germ) (9), the optimisation of protein expression by introducing point mutations without changing the amino acid sequence (10), and Yeast-Two Hybrid (11).
Outlook
Due to the existing clone cooperation with Harvard, we will be able to double the outcome of the subproject as we actively exchange clones and sequence information of our pipelines.
Moreover, we have tested 15 expression vectors for bacteria, yeast, Baculo virus and mammalian protein expression. We offer all Gateway-compatible ORFs for all expression systems (http://www.rzpd.de/products/orfclones/orfclones_2.shtml/#vectorTable). Therefore, RZPD users will have the choice of the expression system and tag used in protein expression. These “ready-to-use” clones have the same quality standards and annotations as the shuttle clones described above.
Lit.: 1. Braun et al., Proteome-scale purification of human proteins from bacteria. PNAS 2002 99, 2654-2659. 2. Braun, P.et al., High throughput protein production for functional proteomics. Nat. Biotechnol. 2003, 21, 383-388.3. Reboul J, et al., C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet 2003, 34:35-41; 4. Gavin AC, et al., Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002, 415:141-147. 5. Liebel, U, et al., A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 2003, 554, 394-398. 6. Lennon, G. et al., The IMAGE consortium : An integrated analysis of genomes and their expression. Genomics 1996, 33, 151-152. 7. Wiemann, S, et al., Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res 2001, 11:422-435. 8. Strausberg RL, et al., The mammalian gene collection. Science 1999, 286:455-457. 9. Langlais, C, et al., Funktionsbiologie benötigt Zugang zu rekombinanten Proteinen: Klonierung, Validierung und Expression, Transkript 2003, 3, 6. 10. Langlais, C, et al., The Linear Template Generation Set: Optimization of Protein Expression in the RTS 100 HY, Biochemica 2003, 3, 22. 11. Stelzl u., et al. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell, 2005, 122, prepublished electronically Sept. 1st.


