Introduction
The post-genomic era has left us with a daunting number of novel proteins of unknown function. For a systematic functional analysis of these proteins, highly parallelised and cost-effective methods are required. Micro-array formats are a proven means of miniaturisation for the analysis of nucleic acids. For proteins, however, the peculiar physico-chemical requirements of the different proteins for activity have severely hampered the use of protein micro-arrays. Here, we present progress on the use of DNA micro-arrays for in situ generation of protein micro arrays. Our method is based on the Nucleic Acid Programmed Protein Array - (NAPPA-) technology (Figure 1). In this method, plasmids directing the expression of GST-fusion proteins are spotted along with an antibody against GST. The fusion roteins produced by use of a coupled transcription/translation reaction are captured and immobilised via their GST tag. Since these arrays are produced just before their use, problems of protein stability can be avoided.
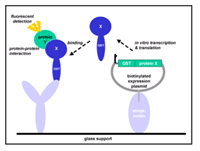
We are interested in two main fields of application of NAPPA-technology: first, these chips are suited for the detection of protein interactions, where an array of potential binding partners are probed with a soluble protein for binding. Secondly, since the proteins on the array should in principle be in a native state, these chips should allow the parallel detection of protein activity. This could be exploited in drug discovery research, where the interaction of a family of drug target proteins with a given candidate therapeutic substance can be examined simultaneously.
Results/Project Status
To quickly establish the basic method at the RZPD, a collaboration with the group of Joshua LaBaer at the Harvard Institute of Proteomics has been established, and the subtleties of the method were learned during a visit to the Boston lab. Conditions were re-examined and titrated at the RZPD. Current activities include
- Use of NAPPA-technology to verify protein-protein interactions predicted by yeast two-hybrid screening
- Expression of proteins on the slide from linear templates generated by PCR
- Detection of protein activity on the slides.
An example for a slide generated by in situ expression can be seen in Figure 2. Currently, we need to gain a better understanding of the factors important for the efficient immobilisation and translation of the DNA, and for the binding of active proteins on the slide. Various expression systems and tags need to be evaluated.

Outlook
The most important potential applications of NAPPA technologies include
- the detection of protein-protein binding,
- the detection of protein binding to small drug-like molecules
- the detection of protein activity
- testing of serum samples or antibodies for their reactivity against human proteins
Ultimately, we believe that the ability to synthesise active protein in a micro-format in combination with systematic collections of DNA constructs for protein expression, such as available at the RZPD, will open up tremendous possibilities in post-genomic research.
Lit.: 1. Ramachandran N et al. Self-assembling protein microarrays. Science. 2004 305:86-90


