Introduction
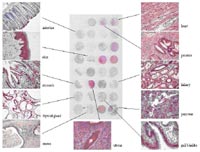
RZPD’s screening service was founded 10 years ago within DHGP. Beeing part of large sequencing projects, the task originally was the isolation of gene fragments from genomic and cDNA libraries. Since then, we have followed new developments in functional genomics. In 2000 we established a gene expression profiling service on RZPD’s whole-genome cDNA arrays from human, mouse, and rat. In meantime, our Affymetrix hybridization group in Berlin takes over most these experiments. The service can fall back on a large portfolio of arrays from human and different model organisms and works on a cost covering basis http://www.rzpd.de/services/affymetrix/). Moreover, the NimbleGen service on highly flexible microarrays allows customized applications (http://www.rzpd.de/services/nimblegen/). Beside our activities in genomics and transcriptomics, we have started proteome analyses on tissue microarrays (fig. 1), RZPD’s protein expression arrays (fig. 2), and antibody arrays (fig. 3) in 2003. According to demands of users within NGFN-2, we currently run different feasibility studies to extend our service portfolio.
Results/Project Status
RZPD’s protein expression arrays (for details see: http://www.rzpd.de/products/proteinarrays/ and report on the project “Infrastructural Basis for Genome Research” by U. Radelof) are widely used for screenings of patient sera or other body fluids (1), characterization of antibodies, or other assays like protein binding studies (2, 3). Screening for auto-antibodies or antibody binding sites has been established during the last year and is now offered as a service (fig. 2, http://www.rzpd.de/services/pr_screening/). For the approach we are now working on, the Human Fetal Brain Protein Expression Array is overlayed with cerebrospinal fluid (CSF) from depression patients (cooperation department C. Turck, MPI for Psychiatry, Munich) to search for autoantibodies which might lead to potential targets for diagnostics and therapeutic applications.
Another issue is spotting of customized antibody sets, because commercial arrays very often depend on certain antibody suppliers and therefore are limited in complexity and choice of model organisms. Based on transcriptome analyses and immunohistochemistry studies in a rat model

on heart hypertrophy (cooperation K. Amann, Univ. of Erlangen), it was possible to select antibodies specific for proteins involved in extracellular matrix remodeling, spot them on glass slides (C. Schmidt, department A. Poustka, DKFZ), and label protein extracts with Cy3 and Cy5 for protein expression profiling. The results of the experiments should allow us to confirm RNA results on protein level (fig. 3). This might be stimulating for other users working on similar projects.
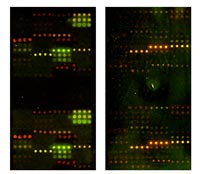
Within our ongoing work on the BD500 antibody array we applied a low sample protocol with protein lysates isolated from formalin fixed tissue slides. And indeed we were able to detect expression differences between normal and tumor tissue with as little as 50µg whole-protein lysate (cooperation K.-F. Becker, TU Munich.). Therefore, this is a very attractive method not only for pathologists, but for all scientists working with microdissections or other small samples.
Outlook
Since our results on antibody arrays and RZPD’s protein expression arrays look very promising, we plan to extend these activities together with NGFN-2 partners. One project will include the test of binding assays on protein expression arrays (RNA, DNA, proteins, sugars, lipids etc.). We will have to test to what extend these methods are appropriate on these arrays containing denatured proteins. As a complementary assay to protein profiling on antibody arrays, we are also planning the reverse assay. This is spotting of proteins extracted from formalin fixed tissue and detection with individual antibodies. This again might be an interesting application for pathologists interested in quantitative detection of individual proteins in low concentrations. Another goal will be functional genome analyses in array format. Whole cells can be spotted onto slides to generate cell arrays (4), which allow high-content screens after transfection with different cDNAs or after siRNA knock-downs.
Lit.: 1. Cepok S et al. Identification of Epstein-Barr virus proteins as putative targets of the immune response in multiple sclerosis. J Clin Invest. 2005 May;115(5):1352-60. 2. Mahlknecht U et al. Far-Western based protein-protein interaction screening of high-density protein filter arrays. J Biotechnol. 2001 Jun 15;88(2):89-94. 3. Lee J et al. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002 Mar;3(3):268-73. 4. Ziauddin J and, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001 May 3;411(6833):107-10.


