Introduction
Integrated genome research requires the costly generation and utilization of large molecular biology resources, e.g. clone libraries, protein expression clones, siRNA resources for the investigation of gene function etc. These are essential resources for biomedical research aiming at the improvement of human health. Today, they already consist of scores of millions of samples and their numbers increase steadily. Maintaining the long-term integrity and safety of this indispensable material, while guaranteeing public access for all researchers, is essential for the National Genome Research Network (NGFN), and molecular biology research in Germany in general. For this purpose, RZPD continuously generates and also imports relevant research material and applies stringent quality controls to assure a consistently high standard. RZPD ensures reliable and unrestricted access to these resources and all material that will be generated and made publicly available from the NGFN for a fee on a cost-recovery basis. These resources and related services are provided on the basis of a reliable, adequate laboratory infrastructure. In order to make sure that the experimental value of the biological material is not degraded over time, RZPD has built a unique system of logistics, maintenance, and quality control procedures. Within RZPD, all production, quality control and validation processes are performed and documented according to ISO9001 standard operating procedures (SOPs).
Focus of Research & Development
As a genuine research service facility, RZPD generally aims at a robust implementation and provision of cutting-edge technologies and resources as well as the collection and integration of related data.
Exploiting our experience in large-scale and high-throughput approaches, we pursue the development of genome-wide resources, e.g. Unigene Collections, Minimal Tiling Path Genomic Clone Sets of entire genomes, Full-length Clone Sets, Full ORF Clones, siRNA Resources, high-content Protein Arrays etc. For the investigation of each gene of a respective genome, RZPD aims at providing at least one validated item of each resource type.
Project Status
Within the last years we have rigorously shifted our focus from providing predominantly uncharacterized reference material, which was essential in the infancy of genome research, to material that is vital in modern molecular biology. Nowadays, resources are requested that are comprehensively annotated and that passed strict verification/validation procedures, e.g. sequence-verification or functional validation.
In order to make it convenient to search all available resources and related information, RZPD developed GenomeCube™ – an intuitive-to-use, powerful search engine exploiting internal as well as external databases and other publicly available information systems (fig. 1).
Fig 1a)
STRONG>Fig 1b)
Besides new powerful bioinformatics tools supporting the exploitation of available resources, a number of new resources and services has been established. Examples are:
1) RZPD Human Genomic Clone Set - Version 1.0
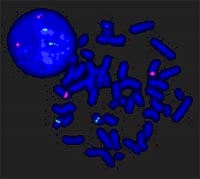
The RZPD Human Genomic Clone Set covers a maximum of the complete human genome sequence with a minimal number of genomic clones. This set is a valuable resource for the functional analysis of genes. Fields of applications are: Fluorescent In Situ Hybridization (FISH) analysis of chromosomal rearrangements (fig. 2), Comparative Genomic Hybridization (Matrix-CGH) analysis of genomic DNA and Protein-DNA binding studies for e.g. promotor analysis.
2) RZPD Protein Arrays
Protein Arrays made of tens of thousands of recombinant proteins derived from a variety of tissues allow for a time-, cost-, and material-efficient functional analysis. With more than 140,000 arrayed proteins, RZPD offers the currently largest collection of arrayed proteins for screening experiments. Examples of applications are: Target protein identification using antibodies/serum/plasma; screening of human serum/plasma for auto-antibodies, epitope mapping of antibodies; functional assays, e.g. phosphorylation, ribosylation, methylation and identification of DNA/RNA binding proteins.
Target protein identification, using antibodies and sera, is also offered as a standard RZPD-service. Protein expression clones as well as sequences from all clones encoding proteins which were identified in a screening experiment are provided upon request.
3) NimbleGen Service Provider
In order to complement RZPD’s Affymetrix’ GenChip™-Service, we introduced NimbleGen’s microarray service. NimbleGen’s technology enables microarrays to be very fast end cost-effectively synthesized for any genome for which quality sequence data is available. RZPD is the exclusive service provider for this technology in Germany and Austria. High-density arrays are currently synthesized with up to 390,000 probes per array. Oligonucleotide-lengths from 24-mers to 70-mers can be chosen.
Current services and applications on NimbleGen microarrays include: Prokaryotic and Eukaryotic Gene Expression Analysis, Chromatin Immunoprecipitation ChIP chip™, comparative genomic hybridization (CGH) and comparative genomic re-sequencing (CGR). Studies with a whole human promoter array will possible very soon.
In summary, based on RZPD’s long-term experience in genome research, its comprehensive resource collection built up during the German Human Genome Project and NGFN-1, a quality management system according to international standards, and the continuous development of new resources for applied genome research according to the requirements of the researchers, RZPD as a non-for-profit Center for Services and Resources” allows for the most economic utilization of restricted money for genome research and all NGFN-2 researchers to benefit as follows:
- Researchers may focus on science and don’t have to invest in the development and maintenance of resources.
- All resources and services are provided with “no strings attached”. All IP rights, which have been generated applying resources form RZPD, belong to the researchers.
- Sustainable provision and distribution of resources developed within DHGP, NGFN or international research projects.
- Centralized quality control and documentation of QC results according to the international standard ISO9001.
Outlook
RZPD’s concept of material and service provision and development, building upon its comprehensive public clone collection and their correlation to the underlying genes covers the entire range from the investigation of the genome to the analysis of phenotypes. Thus, RZPD’s product development always follows the progress in the field and encompasses cutting-edge resources and services for the field of genomics, transcriptomics, proteomics etc. (fig. 3).

Now that the sequences of the human and mouse genome and most of the genes comprised therein are known, the next major challenge in Molecular Life Sciences is to understand the function of these genes, the way they are regulated and expressed, the function of the deduced proteins in the complex biological network, and ultimately the etiology of important diseases, if gene / protein function is not performed correctly. To this end, synthetic resources and in silico approaches have its limitations. Therefore, RZPD’s natural-resource-based approach will certainly remain indispensable. The model of centralization of resource sharing, which allows for the most economic utilization of standardized material of highest quality also supports the RZPD concept.
In 2003, a report of the National Academy of Sciences of the USA (‘Sharing Publication-related Data and Materials: Responsibilities of Authorship in the Life Sciences’) came up with an ethic of data and material sharing: ‘Universal Principal Of Sharing Integral Data Expeditiously’ (‘The UPSIDE of Good Behavior: Make Your Data Freely Available’, Science 2003, 299:990). RZPD’s work actively supports this initiative of open and efficient exchange of material and data for the benefit of the scientific community within NGFN-2 and beyond.
Lit.: 1. Stelzl U et al. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell 2005 Sept; 6(122) 2. Gutjahr C, Murphy D, Lueking A, Koenig A, Janitz M, O'Brien J, Korn B, Horn S, Lehrach H, Cahill DJ. Mouse protein arrays from a TH1 cell cDNA library for antibody screening and serum profiling. Genomics. 2005 Mar;85(3):285-96 3. Ross MT et al. The DNA sequence of the human X chromosome. Nature. 2005 Mar;434(7031):325-37 4. Illiger J, Herwig R, Steinfath M, Przewieslik T, Elge T, Bull Ch, Radelof U, Lehrach H, Janitz M.bEstablishment of T cell-specific and natural killer cell-specific unigene sets: towards high-throughput genomics of leukaemia. Eur J Immunogenet. 2004 Dec;31(6):253-7 5. Kittler R et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004 Dec 23;432(7020):1036-40 6. Weaver T, Maurer J, Hayashizaki Y. Sharing genomes: an integrated approach to funding, managing and distributing genomic clone resources. Nat Rev Genet. 2004 Nov;5(11):861-6 7. Yuille K, Korn B, Moore T, Farmer AA, Carrino J, Prange, C, Hayashizaki Y. The responsibility to share: sharing the responsibility. Genome Res. 2004 Oct; 14(10B): 2015-9.


