MicroRNAs (miRNAs) are single-stranded RNAs (ssRNAs) of 19–25 nucleotides in length that are generated from endogenous hairpin-shaped transcripts in the genomes of plants and animals (refrenced in 1). According to the current knowledge, miRNA genes are transcribed by RNA polymerase II (pol II) to generate the primary transcripts (pri-miRNAs). The initiation step (‘cropping’) is mediated by the Drosha–DGCR8 complex, located in the nucleus. The product of this nuclear processing step is a ~70-nucleotide (nt) pre-miRNA, which possesses a short stem plus a ~2-nucleotide 3’ overhang. Pre-miRNAs are subsequently exported from the nucleus in complex with exportin-5 and Ran-GTP. Following export, the cytoplasmic RNase III Dicer performs the second processing step (‘dicing’) to produce miRNA duplexes. The duplex is separated and usually one strand is selected as the mature ~22-nucleotide miRNA, whereas the other strand is degraded (Fig. 1).
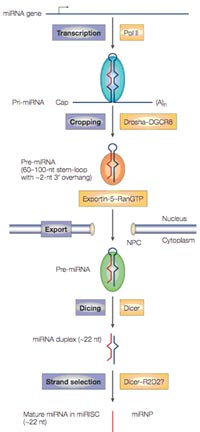
miRNAs appear to function as guide molecules in post-transcriptional gene silencing by base pairing with target mRNAs, which can lead to mRNA cleavage or translational repression. With more than 200 members per species in higher eukaryotes, miRNAs are one of the largest gene families, accounting for ~1% of the genome. miRNAs and their targets seem to form complex regulatory networks, since a single miRNA may bind to and regulate many different mRNA targets and, conversely, several different miRNAs may bind to and control a single mRNA target. A growing body of evidence predicts that more than 30% of all human genes may be targeted by miRNAs. Thus, the unique combination of miRNAs that are expressed in each cell type might affect the utilization of many mRNAs and influence cellular function. Recent studies have provided evidence that miRNAs may act as key regulators of processes as diverse as early development, apoptosis and cell proliferation and cell differentiation. Therefore, miRNA expression is implicated also in disease processes, such as chronic lymphocytic leukemia, breast cancer, colonic adenocarcinoma, viral infections and inflammation. There is speculation that in higher eukaryotes, the role of miRNAs in regulating gene expression might be as important as that of transcription factors and thus, may represent also exceptional novel targets for the development of novel therapeutic approaches (see 2).
Project Status
Although the complexity of this regulatory circuitry is currently overwhelming, one of the first key steps towards dissecting the network would be to understand which miRNAs are transcribed in distinct cell types and how the expression changes with disease processes. Consequently, our project aims to develop a specialized oligonucleotide array to specifically analyze the expression patterns of microRNAs during the development and progression of tumors analyzed by the CancerNet. Our results will not only contribute to the understanding of miRNA regulation and function, but will help to identify corresponding mRNA targets. The established miRNA-array will also be of general value for other disease networks in the NGFN2 and the commercial exploitation of miRNA-based therapies.
In the initial phase of the project, we collected microRNA sequences that have been identified in the human and mouse genome to date.
Oligonucleotides specific for 20 selected microRNAs that either exhibit perfect complementarity to the microRNA sequence or that include mismatches (specificity control) were immobilized on a pilot microRNA array. The array not only contains oligonucleotides representing mature human miRNAs (20-22mer) but also longer oligonucleotides representing pre-miRNAs (40mer). We compared various glass-surfaces and protocols to optimize spotting as well as hybridization protocols. Furthermore, we included positive and negative controls (“spike-in”) in the design of the pilot mi-RNA microarrays as we previously established for conventional cDNA microarrays. This will aid data normalization and background noise calculation during the data evaluation step. Preliminary results indicate that the designed mi-RNA microarray is suitable for the analysis of miRNA expression pattern.
Following the successful application and optimization of microRNA extraction protocols from cells and tissues, we compared the quality and reproducibility of miRNA detection and quantitation using our custom made miRNA-microarrays and Northern blot analysis. As indicated in Fig. 2 we were able to isolate and detect several microRNAs in different organs of the mouse. Importantly, microarray and Northern blot analysis showed a good correlation with regard to sensitivity and specificity of miRNA detection.

Beside the development of improved tools to detect miRNA expression it is also of utmost importance to identify novel proteins involved in tumorigenesis, which might represent disease relevant targets for miRNA mediated gene expression regulation. Consequently, we recently showed in collaboration with the SMP RNA (Sültmann) that the differentiation antigen NY-BR-1 is overexpressed in breast cancer patients and encodes a potential target for antibody based therapies (3).
Outlook
With reliable and sensitive oligonucleotide microarrays that interrogates all known human miRNAs, we will screen for miRNA expression in patient samples suffering from acute lymphoblastic leukemias (ALL), head and neck and breast cancer. It is expected that the differential expression of miRNAs might give insights into prognosis, therapy related toxicity, and the likelihood to develop secondary malignancies. The miRNA expression pattern may also be valuable for the design of risk-adapted individual therapeutic strategies. Furthermore, the identification of miRNA regulated target genes will yield interesting information about the biology of haematopoiesis and malignant transformation.
Lit.: 1. Gong, H., C. M. Liu, D. P. Liu, and C. C. Liang. 2005. The role of small RNAs in human diseases: potential troublemaker and therapeutic tools. Med Res Rev 25:361-81. 2. Kim, V. N. 2005. Small RNAs: classification, biogenesis, and function. Mol Cells 19:1-15. 3. Seil, I., H. Sültmann, S. K. Knauer, E. Jäger, K. Zatloukal, A. Knut, D. Jäger, and R. H. Stauber. 2005. The differentiation antigen NY-BR-1 encodes a potential target for antibody based therapies in breast cancer. submitted.


