Introduction
For a cancer to occur and to progress, apoptosis must be inhibited because the tumor cells are exposed to a variety of constant apoptotic stimuli, i.e. overexpression of apoptosis-inducing oncogenes, hypoxia or the lack of survival signals after detachment of metastatic cells from the tumor mass (anoikis; (1)). Furthermore, conventional treatment of tumors by chemotherapy and irradiation is based on the selective induction of apoptosis in the tumor cells, and whenever a tumor becomes resistant against such treatment, this is achieved by inhibition of drug- or radiation-induced cell death (2).
It has emerged as a promising therapeutic concept to re-introduce apoptosis sensitivity in tumor cells (3). To be able to develop an efficient and clinically relevant strategy it is mandatory to identify all the proteins which are involved in the regulation of apoptosis and which are important mutational targets responsible for apoptosis resistance in cancer.
Two types of apoptosis mutations are observed in tumor cells. Loss-of-function mutations inactivate pro-apoptotic molecules which are needed for the excecution of the apoptotic program. Gain-of-function mutations lead to the often observed overexpression of anti-apoptotic proteins. The prototype of such overexpressed oncoproteins is Bcl-2, the founding member of a new class of oncoproteins which do not support proliferation but rather inhibit apoptotic cell death efficiently. Such anti-apoptotic proteins are attractive targets for inactivation/downregulation in the course of a molecular cancer therapy.
We have successfully established a functional survival screen in the yeast S. pombe which allows to identify new anti-apoptotic mammalian proteins. A pro-apoptotic mammalian „killer“ gene is expressed in S. pombe in an inducible manner which upon induction leads to rapid yeast cell death. By co-transforming a mammalian cDNA library, some of the yeast cells are protected and survive. Such protecting molecules would either inactivate the pro-apoptotic protein by direct binding or by interfering with an endogenous yeast cell death program which is triggered by the killer protein. Screening mammalian cDNA libraries in yeast reduces much of the false positive background experienced when performing such screens in mammalian cells.
Our screen enables functional high-throughput screening of large tumor cDNA libraries. Two different parameters can be adjusted: One is the choice of the „killer“ gene overexpressed in yeast. This decision has impact on the nature of the inhibitory proteins which will be isolated. In addition, a particular tumor type (or therapy-resistant tumor) of interest can be selected as a source for the cDNA library screened in yeast
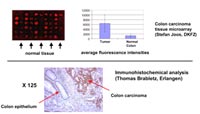
During the first funding period of NGFN 1 we isolated a number of known and unknown anti-apoptotic genes from a breast tumor-derived cDNA library. We could show with the help of several NGFN collaborations (Stefan Joos/Peter Lichter, DKFZ Heidelberg; Thomas Brabletz, Erlangen; Matthias Trust/Christian Hagemeier, Charité Berlin; Geert Michel/Gerhard Przemeck/Martin Hrave de Angelis, GSF München) that the High mobility group 1 protein (HMGB1) is upregulated in human breast and colon carcinomas. These results have been published in two papers (4), (5).
More than 50% of the mammalian cDNAs which inhibited yeast cell death in our functional survival screens were also able to protect mammalian cells against several apoptotic stimuli.
During NGFN2, we are planning to use the yeast survival screen in S. pombe for further systematic large-scale screens of cDNA libraries isolated from different therapy-resistant tumors. Apoptosis resistance in such tumors is likely to be conferred by the overexpression of anti-apoptotic oncogenes which we hope to isolate in our screens. The network of collaborations built up during NGFN1 will be continued in NGFN2 and fed with newly dis-covered anti-apoptotic genes inhibiting tumor treatment
Project Status
We have cloned three new cDNA libraries from different tumor material into a constitutive yeast expression vector. For the first library, several glioblastoma biopsies were pooled. The second library was cloned from a therapy-resistant melanoma metastasis, and the third library was prepared from several different childhood leukemia blasts, some of which were relapses following chemotherapy. Currently we are screening these libraries in our functional yeast survival screen to identify tumor cDNAs which inhibit yeast cell death induced by the Apaf-1 homologue CED4. We expect to find proteins that are able to block mammalian apoptosis downstream of Cyt c release and that may be responsible for tumor resistance against apoptosis-inducing therapy.
Outlook
A postdoc in our laboratory will finish to screen the libraries for tumor cDNAs which allow the yeast to survive Apaf-1/CED4 expression and which inhibit mammalian apoptosis. As a readout system we will use transfection into 293T cells (with high transfection rates) and UV radiation or Apaf-1/Casp-9 transfection as two independent apoptotic stimuli. The identified candidate genes will be subjected to the collaboration network already established during NGFN1. Tools (siRNA, knockout and transgenic mouse models, antibodies) will be generated to analyze the molecular mechanism how these genes inhibit programmed cell death. During these collaborations we will evaluate the expression levels of the isolated genes in different tumor entities. To prove the oncogenic potential of the genes and to test their ability to increase radiation- and chemotherapy-resistance, transgenic overexpression mouse models will be established. Knockout mouse models will help to elucidate the in vivo function of the candidate genes. Such animal models will also be used to evaluate potential targeting strategies which aim at downregulation of the anti-apoptotic genes to re-sensitize tumors for therapeutic treatment. As a tool for proof-of-principle experiments in this context, siRNA (Christian Hagemeier/Matthias Trust, MDC Berlin) will be used.
In summary, we expect to identify and to analyze in a comprehensive way new anti-apoptotic mammalian genes responsible for therapy-resistance of tumors.
Lit.: 1. Zörnig M et al. Apoptosis regulators and their role in tumorigenesis. Biochim Biophys Acta. 2001 Oct 1;1551(2):F1-37. 2. Brown JM et al. The role of apoptosis in cancer development and treatment response. Nature Reviews Cancer. 2005 March;5(3):231-7. 3. Kirkin V et al. The role of Bcl-2 family members in tumorigenesis. Biochimica et Biophysica Acta. 2004 Mar 1;1644(2-3):229-49.4. Brezniceanu ML et al. HMGB1 inhibits cell death in yeast and mammalian cells and is overexpressed in certain human tumours. FASEB J. 2003 Jul; 17(10):1295-7.5. Volp K et al. Increased expression of High Mobility Group Box 1 (HMGB1) is associated with an elevated level of the anti-apoptotic c-IAP2 protein in human colon carcinomas. Gut. 2005 Aug 23;Epub


