
Introduction
In human immunodeficiency virus (HIV-1) -infected patients, the individual genetic background plays a pivotal role for the differential disease progression towards AIDS. The small fraction (5 %) of individuals with almost non-detectable viral loads, stable CD4+ counts and no AIDS-specific disease progression over more than ten years without antiretroviral therapy points out that polymorphic host factors can efficiently suppress retroviral replication. Several factors causing a variable disease course have been identified (1): (i) infection by attenuated viruses with impaired gene functions (eg., vif, vpr, nef) leads to long-term nonprogression (LNTP); (ii) impairment of viral entry by altered cellular host receptors (CCR5, CCR2, CXCL12) delays AIDS; (iii) variations in the major histocompatibility complex (MHC) genes (HLAB*B27, HLAB*B57) are prognostic for slow disease progression (SP) or rapid disease progression (RP); (iii) variant expression of innate immunity molecules (IL10, MIP, RANTES, Kir3DS1) can be involved in lower suscpetibility to HIV-infection. However, despite intensive research, it is estimated that more than 90% of the genes responsible for the development and maintenance of the LTNP phenotype and resistance to HIV infection are undiscovered (1). In addition, the molecular basis of LTNP associated with a soluble factor, termed CD8+ T cell antiviral factor (CAF) which is able to control actively viral replication, remains to be elucidated (2)
Further molecular understanding of resistance to the HIV-induced disease promises insights into natural antiretroviral control mechanisms. However, systematic studies to elucidate this are complicated by the heterogeneity of HIV-infected individuals with respect to genetic diversity, ethnic affiliations, age, gender, disease stage, as well as their individual family and medical history and immunological status prior and post infection. Moreover, time point, viral dose and pathogenicity of the infecting isolate are frequently unknown in human patients.
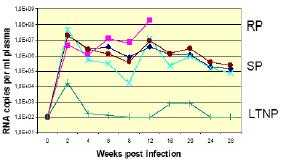 | |
| Fig 1: Viral load during SIV-infection in rhesus monkeys displaying rapid progression (RP), slow progression (SP), and long-term nonprogression (LTNP) of AIDS. | |
To initiate a systematic study on polymorphic host factors especially for LTNP and resistance to HIV infection we refer to the most relevant animal model for the pathogenesis and the treatment of human AIDS (3). We investigate a rhesus macaque cohort bred at the German Primate Center in Göttingen which has been infected under controlled experimental conditions by the simian immunodeficiency virus (SIV). Upon SIV-infection, rhesus monkeys develop pathological and clinical manifestations remarkably similar to those of human AIDS patients, and display a varying disease course (Figure1; RP, SP and LTNP) (3). Since the family history of the cohort members is provided, state-of-the-art genomic screening technology such as quantitative trait loci (QTL) and linkage disequilibrium (LD) mapping allows -for the first time- a genome-wide scan to obtain inheritable susceptibility/resistance markers for AIDS. We have started the combined approach by determining genetic variations (SNP's, InDels, haplotypes) by targeted resequencing of genomic loci of selected candidate genes in SIV-infected rhesus showing variable disease progression. To define accurately MHC-related and non-MHC-related contribution of the disease progression we also extend the molecular typing techniques for the rhesus macaque Mhc genes (4).
The research team includes Ulrike Sauermann and Gerhard Hunsmann from the German Primate Center (Göttingen), Michael Krawczak (Institute for Medical Informatics and Statistics, University of Kiel), Peter Nürnberg (Cologne Center for Genomics, University of Cologne) and Roman A. Siddiqui (Leibniz Institute for Age Research - Fritz-Lipmann-Institute, Jena).
QTL-, LD-mapping
QTL- and LD-mapping is used to identify markers and to map gene loci for inheritable human diseases. To date, there are only few publications on mapping loci controlling the susceptibility to viral diseases. So far no publication dealt with a genome-wide scan for susceptibility markers for AIDS. Here the systematic whole genome scan of SIV-infected monkeys intends to identify and quantify host genome regions that control viral replication, and may reveal novel cellular targets for antiviral strategies.
To achieve an average marker density of 8 cM by the QTL-mapping, the DNA-samples are screened with about 350 different polymorphic STR (short tandem repeat)- markers. To assess the feasibility of the project, simulation based lod score calculations were performed for a subset of monkeys (90 offsprings from 6 families). These results show that the study can be successfully completed.
Resequencing of candidate genes
Here we are determining genomic variability (SNPs, Indels and haplotypes thereof), in a number of genomic loci of candidate genes in SIV-infected rhesus representing the extreme ends of disease (RP, LNTP). As an entry we are characterizing the pathogen receptors DC-Sign (5) and Toll (6) , and the defensins (DEF; 7) which have been shown to display anti-HIV activity (8). At the current state of analysis in three LTNP, seven RP samples, and in genomic DNA of an uninfected rhesus, as a control we detected polymorphisms potentially playing a role in the course of SIV-infection. Several indels and single base variations in the genomic rhesus DC-Sign region (CD209, CD209L2) ) are corresponding within RP and LTNP samples, respectively. Preliminary characterization of the Toll-like receptor genes (Tlr3, 4, 7, 9) indicates less genomic variations between RP and LTNP rhesus monkeys. DC-Sign acts as receptor for HIV and SIV on macrophages and dendritic cells, and is able to enhance the infectivity of immunodeficiency viruses (5). Recently, the Toll-like receptors have been hypothesized to be involved in the variable susceptibility towards opportunistic infections in RP, SP and LTNP (6).
Genotyping of rhesus MHC-class I alleles
To evaluate the contribution of Mhc class I haplotypes to the highly variable disease course in SIV-infected rhesus macaques, we determined Mhc class I haplotypes by sequencing about 1800 Mhc class I cDNA clones from pedigreed monkeys. Since macaques have multiple A and B genes, we detected per haplotype up to 3 A-genes and up to 7 expressed B-genes. We are are currently developing rapid typing techniques for 38 Mhc class I alleles in order to validate the haplotypes in a larger number of macaques. So far, evidence suggests that the general disease course is determined by Mhc class I genes in most animals. However, the status of LTNP cannot be explained by presence of Mhc class I genes alone.
Outlook
The combined approach to determine polymorphisms by targeted resequencing of QTL-gene/genomic loci, and of selected candidate genes, allows to correlate variant host factors especially to the extreme ends of AIDS, RP and LTNP with help of the AIDS Macaque model.
Most importantly, the results obtained from the animal model will be used for the inspection of a human HIV-infected patient cohort, mediated by the “Kompetenznetz HIV/AIDS” (www.kompetenznetz-hiv.de).
The identified loci, the genetic variations in structural genes and regulatory determinants, will be deposited in a www-accessible database facilitating functional and mechanistic studies by the research community.
Moreover, health-related research on nonhuman primates is conducted in more than 30 European institutions supported by private or public funding, with increasing demand of genetically characterized monkeys. Here, the identification of novel markers predisposing to a specific disease pattern will allow to pre-select monkeys prior to the experiment and thereby significantly improve the quality of the results. Finally, employing genetically defined animals will reduce their overall number required for individual preclinical trials.
The recognition of specific genetic patterns relevant for susceptibility to infection and disease will ultimately assist us to improve the diagnosis, prognosis, and therapeutic interventions of human AIDS patients.
Lit.: 1. O'Brien SJ, Nelson GW. 2004. Human genes that limit AIDS. Nat Genet. 6:565-74. 2. DeVico AL, Gallo RC. 2004. Control of HIV-1 infection by soluble factors of the immune response. Nat Rev Microbiol. 2:401-13. 3. Haigwood NL. 2004. Predictive value of primate models for AIDS. AIDS Rev. 6:187-98. 4. Mühl T, Krawczak M, Ten Haaft P, Hunsmann G, Sauermann U. 2002. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J Immunol. 169:3438-46. 5. van Kooyk Y, Geijtenbeek TB. 2003. DC-SIGN: escape mechanism for pathogens.Nat Rev Immunol. 3:697-709. 6. Bafica A, Scanga CA, Schito M, Chaussabel D, Sher A. 2004. Influence of coinfecting pathogens on HIV expression: evidence for a role of Toll-like receptors. J Immunol. 172:7229-34. 7. Taudien S, Galgoczy P, Huse K, et al. 2004. Polymorphic segmental duplications at 8p23.1 challenge the determination of individual defensin gene repertoires and the assembly of a contiguous human reference sequence. BMC Genomics 5:92-9. 8. Lehrer RI. 2004. Primate defensins. Nat Rev Microbiol 2:727-38


