Introduction
Next to recombinant proteins, synthetic peptides will be the second pillar on which the SMP "antibody factory" builds its antibody selection technology. We expect that a significant part of the chosen protein antigens will not be produced in sufficient quality and quantity by high-throughput recombinant expression. For these antigens we will provide peptide fragments taken from the primary amino acid
sequence. GBF methodology will be applied for the selection of the peptides and their chemical synthesis. Additionally, synthetic peptides will have an important function in the validation process of the antibody factory: every delivered antibody will be accompanied by detailed information on its specificity acquired also through peptide-based epitope analysis.
Project Status
We have been active in the field of peptide synthesis and immunological applications since 1985. Besides research on improving methods and approaches concerning chemical peptide synthesis which are worldwide recognized and practiced in academic and industrial settings, a potent GBFinternal central peptide synthesis and screening service is operated which produces over 200 large-scale peptide
preparations and almost 30000 peptides on arrays per year. This assures an infrastructure adequate for the goals of this SMP project.
Work package 1: Selection of peptide antigens
Within a joint project on “Development of Platform Technologies for Functional Proteome Analysis – Application to the Human Brain”, a project of NGFN1´s Proteomics Technology Platform, we have developed an approach for the automated genome-wide mapping of protein-protein interactions based on the use of peptide arrays [1]. Our experimental approach utilises a human brain cDNA library
expressed and presented on bacteriophages. These are affinity enriched (panned) on synthetic peptide ligands derived from brain specific protein sequences and arrayed on a plastic surface. In the course of this project work we have implemented a new software tool, the “Accessible- Surface-Scanner (ASS)“, which takes listings of protein sequences by data base accession numbers such as from TREMBLE, scans the protein sequences for peptide fragments that have a higher propensity of being located on the surface of the folded protein, thus, accessible for interaction with binding partners and puts these into a
listing for the SPOT peptide synthesizer. With this software tool the number of peptides needed for a complete protein scan can be reduced to about 50%. During the project work of the antibody factory, the selection parameters will be carefully validated and optimized.

Work package 2: Peptide production
This part of our project contributions is concerned with providing a professional service for the rapid generation of peptide fragments to be used in phage panning and antibody characterization. For this, our well established peptide SPOT-synthesis will be applied (Fig. 3). The peptides will be chemically assembled on a cellulose membrane support in an array format suited to the scale required for the panning experiments [2]. 5 nmol of peptide can be recovered from a SPOT of a 384 array format. The peptides will be furnished
with a biotin tag at their N-terminus so that they can be reimmobilized to a panning surface via established streptavidin technology. In this format they will be handed over for phage panning.

Work package 3: Epitope analysis
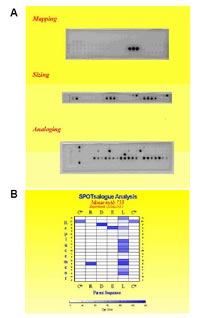
The SPOT peptide array method will be applied also to fine characterize specificities of the antibodies that have been obtained from panning. A typical series of analyses is shown in Figure 4A. Mapping with a complete set of overlapping peptide fragments derived from the protein antigen identifies the epitope region (not required for antibodies obtained from peptide antigens). Then, N- and C-terminal truncations define the minimal epitope sequence (sizing). Systematic replacement with all twenty proteogenic amino acids at all
positions in the minimal epitope (analoging) will give a comprehensive picture of the specificity of the antibody (Fig.4B). This will then allow to search genomic sequence data for potential cross reactivity. An antibody together with this detailed information is a very valuable tool.
Work package 3: Selection of antibody pairs for “sandwich ELISA”
The above described process for epitope analysis can be also applied to polyclonal pools revealing several reactive peptide regions on a mapping array. These reactive peptide regions can be re-synthesized in an immobilized format on a chromatography resin and be used for preparative isolation of the peptide specific (mono-specific) antibody from the pool [3].
A spot on a peptide membrane can capture substantial amounts of antibody. This can be recovered after separating the spots (cut or punched out) and eluting under mild dissociating conditions such as pH 2.5. For good binding peptide spots, the antibody material eluted is sufficient for subsequent analyses such as testing all binary combinations of a series of fractions obtained from a polyclonal pool for
suitability in an ELISA sandwich assay. Peptide epitopes of the best pairing can be synthesised as above for chromatographic purification.
Work package 4: Process for automated multiplexed panning
In order to develop this whole peptide based panning process to full automation, a new array surface will be explored which will allow peptide-addressed elution of the phage libraries directly from the array (Fig. 5). This new peptide array surface avoids the physical separation (cutting or punching) of individual peptide sites from the array before elution of phages as it is required when using a continuous porous cellulose membrane. By lithographic structuring and chemical activation of a hydrophobic plastic foil, an array of
small patches of hydrophilic resin is grafted. The remaining hydrophobic zones separate the patches which can now be individually solvated during the synthesis steps with reagent solutions as well as during the dissociation and elution of bound phages with e.g. detergent solution. There is no risk of cross-contamination between neighbouring patches. This is an important prerequisite for the automation of the process.
We need to systematically explore the production parameters of this support in order to obtain best results in the phage panning experiments. So far, peptide spots on cellulose perform more efficiently by a factor of 100 when comparing the enrichment in model studies with T7 phage display of protein modules. Nothing is known yet about the behaviour of antibody-fragments presenting filamentous phages.

Outlook
Our contribution to the goals of the antibody factory is primarily to provide synthetic peptide technologies. Methodological developments include i) bioinformatic supported prediction of peptide antigens by validating a recently developed software module, ii) isolation of pairs of complementary antibodies suitable for a sandwich-type of ELISA, and iii) optimisation of a new type of peptide array prepared on a hydrophilic structured hydrophobic plastic surface. A significant improvement of through-put in the peptide-based phage affinity enrichment is expected by adopting the patch-peptide array to the multiplexed panning with antibody-fragments presenting filamentous phages.
Lit.: 1. Bialek K et al. Epitope-targeted proteome analysis: Towards a large-scale automated protein-protein-interaction mapping utilizing synthetic peptide arrays. Anal. Bioanal Chem. 2003;376:1006-1013. 2. Frank R. Spot-Synthesis: An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217-9232. 3. Rübenhagen R. Ein neuer Weg zu Antikörper-Mikroarrays mit höherer Spezifität und Selektivität. BioSpectrum. 2005; in press.


