
Introduction
RNA interference (RNAi) technology has been implemented in mammals to study gene function in tissue culture, and, more recently, also in vivo. The major promises of this technology are that it is less time-consuming as compared to homologous recombination in ES cells, allowing fast evaluation of gene functions in vivo. It also permits the inactivation of several genes simultaneously by co-infection or co-expression of several independent RNAi species. Furthermore, in combination with conventional null and conditional mutants generated by homologous recombination, complex genetic interactions can be dissected to model quantitatively polygenic traits. In the future, many candidate genes for human diseases will be identified based on positional cloning and comparative gene expression studies of patient material and of pharmacologically treated animals using microarray analysis. In vivo validation of disease candidate genes is presently a major bottleneck for disease gene annotation. The RNAi in vivo technology promises to speed up this process. The major objective of this subproject is to use the methods developed in subproject 4 to implement RNAi technology in vivo to model complex human diseases. Towards this goal we will first focus on animal models of various diseases represented in the disease-oriented genome networks (KG) of the NGFN. We will focus on the functional analysis of candidate genes for Parkinsonís disease (in collaboration with Prof. Gasser and Prof. Riess / University of Tübingen) and for depression (in collaboration with Prof. Holsboer / MPI for Psychiatry, Munich). Thus, this subproject is of high medical relevance and we envisage that additional studies will follow after successful implementation of this in vivo validation pipeline.
Project Status
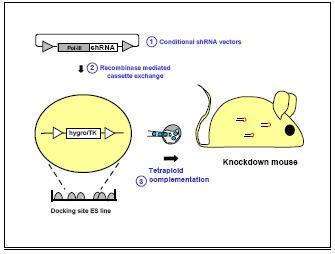
Fig 1: Construction of shRNA vector transgenic mice through recombinase mediated insertion into ES cells.
Validation of siRNA technology to establish animal models for neurological and psychiatric diseases.
To validate RNAi technology in vivo we will compare the RNAi knock-down phenotype with the corticotropin-releasing hormone receptor 1 (CRH-R1) null mutation and conditional mutation which we have established in our laboratory (1, 2). Conditional inactivation of CRH-R1 in the limbic system leads to reduced anxiety-like behaviour. Therefore, we will inject naked siRNA as well as lentivirus containing siRNA of CRH-R1 into the amygdala and hippocampus of mouse brains. Furthermore, we will conditionally induce CRH-R1 specific siRNA in the limbic system using transgenic techniques, followed by the comparison of the mutant phenotype with the established knockout models. Depression is a multifactorial disease in which several loci of chromosomes 2, 4, 8, 11, 12 and 18 are believed to be causally involved. In addition, based on anti-depressant action it has been shown that serotonin, dopamine, noradrenalin and corticotropin neurotransmission is involved in disease aetiology. Based on association studies of patients with major depression SNPs in FKBP5, a cochaperone, have been identified. First, we will knock-down FKBP5 to study whether it is involved in the disease phenotype by injecting naked siRNA or lentiviral vectors containing siRNA into the mouse limbic system. Animals will be analysed with respect to anxiety-like behaviour and neuroendocrine status. Morbus Parkinson is, genetically, a very heterogeneous disease. To date, eleven loci have been associated with Parkinsonism, five of which have been cloned (Parkin, a- synuclein, DJ-1, Nurr1, UCH-L1, Pink1 and LRRK2 (Park8) (3).
Mutations in parkin, a-synuclein, LRRK2 and DJ-1, represent the most common genetic cause of Morbus Parkinson. The significance with respect to the disease phenotype is still unclear for Pink1 and LRRK2 and needs to be validated. Animals will be tested by histology, immunohistochemistry, electrophysiology and behavioural analysis. Presently, we started the project with the construction of transgenic knockdown mouse strains for CRH-R1 and LRRK2 (Fig. 1; see also SMP RNAi project on in vivo technology), we established the stereotaxic injection into the amygdala and build up behavioural assays to test for anxiety and depression in mice. Next, we will establish the production of viral vectors containing shRNA knockdown constructs.
Outlook
We will establish RNAi as an alternative to the use of knockout technology to characterise gene functions in the entire organism. At present we are in the process to assemble the various tools required and expect first results early in 2006. We will provide a platform for in vivo analysis of disease genes identified by the clinical networks within NGFN-2.
Lit.: Timpl P. Impaired stress response and reduced anxiety in mice lacking a functional corticotropin-releasing hormone receptor 1. Nat. Genet. 1998 19:162-166. 2. Müller MB et al. Limbic corticotropin-releasing hormone receptor 1 mediates anxiety-related behaviour and hormonal adaptation to stress. Nat. Neurosci. 2003 6:1100-1107. 3. Krüger R et al. Parkinsonís disease: one biochemical pathway to fit all genes? Trends Mol Med. 2002 8:236-24.


