
Introduction
The diversity of the human genome is a decisive factor in the causation and pathogenesis of complex disease as well as for the individual response to biologically active substances (pharmacogenomics). A key step in all strategies for disease gene identification is the comparative sequence analysis of candidate genes in patients and controls to identify those specific sequence varia-tions associated with complex disease. The two laboratories involved in this subproject (MPI¬MG, Berlin and FLI, Jena) have installed a roboter based automated high-throughput re-sequencing service platform open to all members, cooperating in NGFN (Fig.1, Fig. 2).
This plat¬form is specifically designed as a disease identifica-tion pipeline, to test hypotheses about the involvement of specific genes in inherited diseases, or to verify candidate genes identified by positional or functional cloning projects. 1111

Fig 1: Robotics for Automated Re-sequencing.
We therefore have also implemented bioinformatic techniques (programs to predict haplotypes and haplotype block struc-tures) to analyse complex genotype-phenotype relations. The need for such a service infrastructure has already been ex-pressed by a number of researchers with a strong interest in candidate gene sequencing and gene-based haplotype analy-sis from the various clinical networks.
The coordination of the various sequencing requests from the network partners will be performed through our WEB site (http://www.resequencing.mpg.de/). This information is at all accessible to our partners and it should be obvious to all, what will be sequenced, what has been sequenced so far and who is the responsible person for the specific project.
Project Status
Sequencing Projects
Currently, we are sequencing different candidate genes for complex diseases or pharmacologically relevant genes in cooperation with our clinical partners. A successful project in the past was the analysis of the DNAH5 gene in indiviuals with primary ciliary dyskinesia (PCD). PCD is characterized by recurrent infections of the respiratory tract due to reduced mucociliary clearance and by sperm immobility. Half of the affected offspring have situs inversus (reversed organs), which results from randomization of the left-right (LR) asymetry (Fig. 3). Sequence analysis in individuals with PCD with randomization of LR asymetry iden-tified mutations resulting in non-functional DNAH5 proteins (Olbrich et al., 2002).
Presently, we are analysing the whole DNAH5 gene (77 ex-ons, 14 kb cds) in 94 individuals to identify the complete poly-morphic spectrum of the gene. Until now we have found a very high genetic variability in this region, with a total of 132 SNPs and 12 indels. To indentify interactions of these variations we also use different algorithms to estimate haplotypes and hap-lotype block structures of the DNAH5 gene (Fig. 4.).
Other projects at FLI were focussing on the genetic predispo-sition for chronic inflammatory diseases (Crohn's disease, sarcoidosis) and obesity. Detailed analysis were done for the candidate genes CARD15, BTNL2, MCHR1, MC4R, ghrelin and a gene desert on chromosome 16. Presently, the complex SNP-related sequence variation in the segmental duplicated beta-defensin cluster on 8p23 is determined with respect to inflammatory disorders.
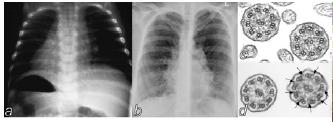
Fig 2: Robotics for Sample Preparation
Presently we are working to enhance our quality management system for the whole re-sequencing process. The system integrates all steps in re-sequencing - from primer design, PCR amplification and cycle sequencing reactions, to capillary electrophoresis systems on the 3730xl DNA analyser and the data analysis. For all these steps we have established stan-dard operating procedures (SOPs).
Outlook
High-throughput re-sequencing of PCR products is the method of choice to identify unknown polymorphism. We offer our high-¬throughput re-sequencing service platform to all mem-bers of the NGFN, to analyse candidate genes or genomic regions of interest.
Using nested PCR assays under stringent conditions, resulting amplicons up to 600 bp can be sequenced in both directions, to detect single nucleotide exchanges, insertions and dele-tions as well as micro-satellites and VNTRs.
Template preparation, purification and modification will be done using robotic systems developed for HT genomic se-quencing projects. The number of patients and controls to be sequenced will depend on the allele frequency to be detected. Maximum numbers will range from 48 up to about 250 sam-ples (alleles with an 1 % frequency). The data will be made accessible to the clinical partners (Fig. 5) and after detailed analysis will be made publicly available.
The Coordinator of the subproject is responsible for quality control of samples and communication with partners. The cost for this service pipeline was calculated to 0.045 Euro per se-quenced base/sample, to be financed by the user requesting the service. This price included selection of primers, PCR amplification, sequencing reactions and data analysis. If exon-intron amplicons are already available, it is possible to send in the PCR products and primer for sequencing and data analy-sis (4.40 € per sequencing read). In this case we will work with the PCR products of our cooperation partners.

Fig 3: a, Half of the PCD-patients show a complete situs in-versus. b, Bronchiectasis. c, Electron micrograph of cross-sections of respiratory cilia from an affected individual of PCD-family. Absence of outer dynein arms is observed on all pe-ripheral doublets whereas healthy individuals d, show normal dynein arms.


