
Introduction
Genetic instability is a hallmark of neoplasia and is thought to be instrumental in cancer initiation and progression. Numerous techniques have been developed for the mapping of chromosomal aberrations, but the identification of candidate genes has been hindered by their low resolution and limited throughput capacity. A possible solution to this predicament emerged with the development of matrix-CGH, a chip-based method for the genome-wide profiling of DNAcopy number status. In matrix-CGH, DNA from a test (e.g., tumor) and a reference genome (genomic DNA from a normal individual) are differentially labeled and hybridized to an array of large-insert genomic clones of known map position. The fluorescence ratios of individual clones provide information about the relative copy number of the corresponding sequences in the test and reference genomes. The resolution and sensitivity of matrix-CGH are determined by the size and mapping position of the clones on the array.
Application of genome-wide matrix-CGH in different tumors is revealing that selection for specific genes relevant to tumor pathogenesis as well as effects of specific types of genetic instability shape tumor genomic profiles. This causal, nonrandom relationship of genomic profile and tumor pathomechanism offers a unique opportunity to address important clinical questions such as differentiation of tumor subgroups with different clinical behavior in regard to prognosis, probability of progression, and therapy response. Furthermore, recurrent deletions or amplifications of genomic material usually are good indicators of the presence of tumor suppressor or oncogenes in the imbalanced chromosomal region. Fine-mapping of the affected regions using highresolution matrix-CGH arrays will facilitate the identification of new target genes for therapeutic interference, especially when combined with the analysis of DNA methylation status, gene- and protein expression.
Well-known examples of diagnosis and treatment decisions based on analysis of genetic aberrations include Herceptin therapy of breast cancer patients with ERBB2 amplification, and the use of STI 571 (Glivec) for tumors with Bcr-Abl translocation or c-Kit mutation. It seems likely that many more such markers will find entry into clinical applications in the future. Genomic DNA has high chemical stability, and its copy number does not vary over time or under different physiological conditions. This makes centralized and standardized DNA-based processing and diagnosis of tumor biopsies a realistic option, in particular since molecular methods to handle genomic DNA already are well established in many clinical laboratories.
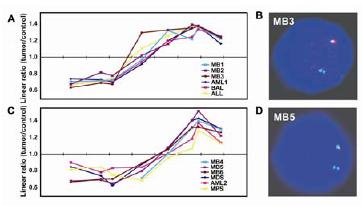
Fig 1: Isochromosome 17 breakpoints in chromosomal band 17p11.2 in medulloblastoma. Two breakpoint clusters (panels A and C) were identified by matrix-CGH and independently confirmed by FISH (panels B and D).
We have completed the construction of a matrix-CGH chip designed specifically for oncogenomic DNA copy number screening. It is based on a 6000 clone chip that we developed within the framework of NGFN1. The chip consists of more than 8000 large-insert genomic DNA clones, mostly BAC (bacterial artificial chromosome), and some PAC (P1-element derived artificial chromosomes) and cosmid clones. 3200 of these clones were selected to map at regular intervals throughout the genome and provide a minimum genome-wide resolution of about 1 megabase (Mb). Another 1800 clones were selected to cover known oncogenes and tumor suppressor genes, as well as genes of potential oncogenic relevance such as key regulators of signaling pathways. The remaining 3000 clones were selected to provide high-resolution coverage of chromosomal regions that frequently are imbalanced in tumors and regions that are gene rich. These include 1p34-36, 1q21-32, 13q14, 13q33-34, 17p11.2, 19, 22q, and a number of smaller regions. With this chip we can concurrently (i) screen tumor genomes for DNA copy number imbalances at 1 Mb or better resolution genome-wide, (ii) monitor the DNA-copy number status of cancer-relevant genes, and (iii) fine-map chromosomal aberrations in selected chromosomal regions. Currently we are providing this chip to seven research groups, several of which are participating in NGFN2. In cooperation with our clinical partners, furthermore we are conducting screening studies of different tumor entities at the DKFZ. These include multiple myeloma (in co-operation with Prof. Hartmut Döhner at the University of Ulm), oesophagal adenocarcinoma (Prof. Martin Werner, Univ. Freiburg), gliomas and ependymoma (NGFN2 Brain Tumor Network). In recently completed studies on medulloblastoma, we identified CDK6 as a novel independent prognostic marker (1), and could map the breakpoints involved in isochromosome 17 formation (Figure 1) with the help of the 17p11.2 tiling-path clones included on the matrix-CGH chip (2)
Outlook
Our work on medulloblastoma and ongoing screening studies of other tumor entities demonstrate that matrix-CGH is a powerful technique capable of identifying clinically relevant molecular markers and putative therapy target genes. The potential of the method can further be enhanced by parallel and comparative profiling of DNA methylation, RNA and protein expression. In co-operation with our partners within and outside NGFN2 we will continue to pursue this type of integrative approach to cancer tumor research.
Lit.: 1. Mendrzyk F et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. JClinOncology 2005, in revision. 2. Mendrzyk F et al. The isochromosome breakpoints on 17p in medulloblastoma are flanked by different classes of DNA sequence repeats. submitted.


