
Introduction
Since the mid 1990s many research groups have focused on the analysis of individual promoters of genes (Kornberg, 1996, Novina and Roy 1996, Roeder 1996, Verrijzer and Tjian, 1996, Ptashne and Gann, 1997, Smale, 1997), to determine the function of even individual bases within the promoter elements for the transcription machinery. But only since the availability of the complete human draft sequence (The International Human Genome Mapping Consortium, 2001) and the advent of large-scale gene expression profiling data on the genome-wide basis, has made possible the ability to correlate the transcript expression levels with the promoter elements of the respective genes. (Ren et al., 2000; Iyer et al., 2001, Pugh and Gilmour 2001). This research has just been started, and it will be essential to provide cloned material for as many promoters as possible to allow functional studies on these elements. Since complete genome sequences are more and more available, bioinformatic tools become appropriate to identify gene specific regions, which are functional in character, e.g. protein encoding or regulatory and are suitable for large-scale complex functional genomics. In this project, we envisage to build a resource of human promoter regions that are cloned into appropriate reporter constructs for complex functional genomics as described for example in the SMP DNA project: "Functional Promoter Analysis", of for RNAi, tissue arrays, functional characterization of promoter elements, promoter/protein interactions, etc. This resource is based on the currently available sequence information of promoter regions, which is continuously updated during the project in the SMP DNA project "Promotor Informatics"‚ and may rise up to 18.000 predicted gene specific promoters. The SMP DNA project "Promoter Resource" is based on the sequence information available for promoter regions identified in SMP DNA project "Promoter Informatics" and directly provides the basis for the systematic functional analysis of promoter regions SMP DNA "Functional Promoter Analysis" (outlined in Fig 1.) as well as other SMPs or the analysis of regulatory networks of candidate genes in KGs. As mentioned in the project "Promoter Informatics" only a small number of promoters are mapped experimentally. Today many theoretical as well as experimental approaches are going on to identify conserved regulatory regions. Currently more then 10,000 promoter regions have been predicted and 6 - 8,000 will be added during the funding period. In the NGFN-1 optimization fund, the RZPD established a set of clones representing a minimal tiling path for the human genome, which will be used as templates for the amplification of promoter regions. This Human Minimum Tiling Path Set – RZPD1, comprises about 25,000 clones, nearly all of them are PAC and BAC clones which were used within the international effort to sequence the human genome. Therefore the insert sequence of the vast majority of the clones has been determined experimentally. This is the only publicly available set that is at least partially verified by a third party. Technically, subsequently promoter regions identified in the SMP DNA project "Promoter Informatics" will be PCR amplified using gene specific primers and cloned upstream of a reporter gene (GFP) using Gateway based systems. The RZPD and MPIMG have the technology and experience in handling large-scale amplification reactions and subsequent cloning. The necessary infrastructure at the RZPD for clone handling and distribution is established and has been successful over the past years. The generated library of cloned promoter regions will be used for different functional genomic approaches as mentioned above and will be available for all NGFN-2 partners.
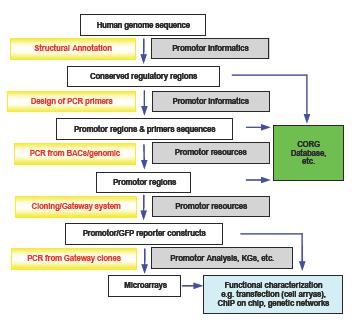
Fig 1:An overview of the experimental outline and the direct interaction with SMP DNA projects "Promotor Informatics" and "Functional Promoter Analysis" is indicated.
The main goal of this project is the generation of about 3,000 human promoter reporter constructs as a resource for functional genome analysis. Our strategy is, to amplify regions of about 1 - 2kb upstream from the start-codon promoter regions, where the transcriptional start sites (TSS) have been experimentally determined both in man and in mouse, and where the orthologous promoter regions are known (Set A; SMP DNA project "Promoter Informatics").
By using gene specific primer sets promoter regions will by amplified and subsequently cloned into a shuttle vector system (Gateway™) that allows the combination of any promoter with any gene of interest. The primer design and template (BAC clone) identification is part of the SMP DNA project "Promoter Informatics". The amplification and cloning of these constructs will be done in close collaboration with the RZPD. The RZPD has well established the infrastructure for large-scale PCR, cloning, clone handling, and clone distribution, including the necessary quality control. As mentioned before, the RZPD has established a Minimum Tiling Path set for the human genome, which will be used as templates for the promoter amplification. All promoter-reporter clones will be sequence-verified at the MPI-MG and constructs will be randomly tested in transfection assays. The promoter-reporter clone resources will be directly provided to the SMP DNA project "Functional Promoter Analysis" for a systematic functional characterization by transfected cell arrays and will be available for analyzing functional regulatory networks within other SMPs or KGs. All data generated will be deposited into public databases (e.g. DBTSS, CORG, EMBL Bank etc.) and the resources will be available in a non-discriminative way to all members of NGFN-2 via the RZPD. This project started very recently and currently we have started to optimize the PCR condition of a test-set of promoter regions.
Outlook
The amplification and cloning of human promoter regions represents the development of a key resource for functional studies on transcriptional regulation. We envisage to clone about 3,000 promoter regions, where the transcriptional start sites (TSS) have been experimentally determined both in man and in mouse, and where the orthologous promoter regions are known. Beside amplification of about 3,000 promoter regions, these amplicons will be cloned into a reporter vector system (Gateway™) that allows the combination of any promoter with any gene of interest. Therefore this resource will serve at least to two distinct approaches: (i) individual analysis of promoter function, and (ii) chip approaches for in vitro promoter binding studies of transcription factors, e.g. cell arrays or DNA microarrays. Both lines of research are followed up in NGFN-2 and are cutting edge technologies to assay gene-function relationships. Pilot amplification experiments have shown that human genomic DNA cannot be used for automated amplification of large numbers of human promoters. In contrast, recombinant genomic clones (cosmids, PACs, BACs) harboring only 40-300kb (more than 104 times reduced complexity of the template) are much better substrates in the PCR process to amplify 1-2 kb of the upstream region of each human gene. In order to obtain a most defined region of genomic DNA, we propose to include the first exon in each amplicon produced if possible. All data generated will be deposited into public databases (DBTSS, CORG, and EMBL Bank) and the resources will be available in a non-discriminative way to all members of NGFN-2 via the RZPD.
Lit.: 1. Kornberg RD. RNA polymerase II transcription control. Trends Biochem Sci. 1996 Sep;21(9):325-6. 2. Novina CD & Roy AL. Core promoters and transcriptional control. Trends Genet. 1996 Sep;12(9):351-5. 3. Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996 Sep;21(9):327-35. 4. Verrijzer CP & Tjian R. TAFs mediate transcriptional activation and promoter selectivity.Trends Biochem Sci. 1996 Sep;21(9):338-42. 5. Ptashne M & Gann A. Transcriptional activation by recruitment. Nature. 1997 Apr 10;386(6625):569-77. 6. Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997 Mar 20;1351(1-2):73-88. 7. Ren et al. Genome-wide location and function of DNA binding proteins. Science. 2000 Dec 22;290(5500):2306-9. 7. Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001 Jan 25;409(6819):533-8. 8. Pugh BF & Gilmour DS. Genome-wide analysis of protein-DNA interactions in living cells.Genome Biol. 2001;2(4):REVIEWS1013. Epub 2001 Apr 4.


