
Introduction
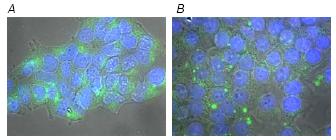
Fig 1: Hek293T cells were transfected with Alexa Fluor 488-labeled negative siRNA (Qiagen) using HiPerFect (A) and Lipofectamine 2000 (B) Transfection Reagent. Images were taken 48 hours after transfection. Alexa Fluor 488-labeled siRNA is green. Nuclei are stained with DAPI.
The aim of this project is to gain insights into the nature and complexity of regulatory networks controlling eukaryotic gene expression at the level of the transcription.
We plan to systematically knock down the expression of ca. 150 human transcription factors in a panel of cell lines via RNAi technique and to analyse the downstream effects of each knocked-down transcription factor on its targeted gene product(s), looking at the global cell transcriptome by means of microarrays.
We will focus our study on transcription factors that are potentially involved in human pathologies and developmental processes, in particular those playing a role in brain development. We also plan to analyse the effect of knocking down the transcription factors encoded by human chr.21 as well as transcription factors known (or predicted) to regulate the expression of key chr.21 genes. This will give insights on regulation networks that may contribute to the pathophysiology of trisomy 21 or Down syndrome, but will also contribute significantly to our understanding of gene regulation networks in general, since the expression pattern of genes on chromosome 21 is very well studied and the analysis of the promoters of chromosome 21 genes is currently ongoing in a number of functional genomics pipelines. RNAi molecules will be designed, tested on cell lines, and the actual gene knock down efficiency will be checked at various time points post transfection by real-time RT-PCR using mock transfected cells as controls. RNAs will be isolated, labelled and analysed by microarrays, and data analysis will be carried out using already established algorithms. We will also integrate all levels of information already available for the selected transcription factors and their targets (e.g. expression patterns of orthologous genes, mutant phenotypes, etc.). With the data generated here, we hope to get a better understanding on associated transcriptional regulation network and use this information to model some relevant transcriptional regulation pathways.
Project Status
Since our sub-project started in Mai 2005, we are still in the process of completing the initial phases of optimization and selection.
1 - Selection of transcription factors
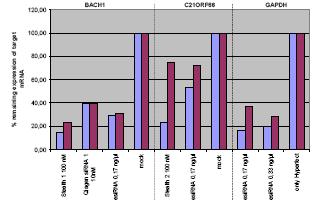
Fig 2: Hek293T tranfections with the indicated siRNAs and esiRNA were carried out using HiPerFect and Lipofectamine 2000 in 12 well plates. Target mRNA levels were measured by qRT-PCR, determining delta-Ct values normalized against ß actin (ACTB) from samples harvested 48 hr post transfection.
Currently, there is no public database allowing the easy retrieval of an exhaustive list of human known and putative transcription factors (and their mouse orthologs) associated with annotation pertaining functional information. The commercial TRANSFAC database (http://www.gene-regulation.com/) is one of the most specific data source containing ~1200 annotated human transcription factors but it is not exhaustive and does not feature putative transcription factors.
We collected a non-redundant list of human and mouse transcription factors by exploiting TRANSFAC information and annotation available in public databases such as SMART, ENSEMBL (GO annotation), CELERA, and WashU (hg.wustl.edu) for the human as well as TRANSFAC, SMART, ENSEMBL (GO annotation) and MGI (Jackson lab) for the mouse. The sets of sequences corresponding to each of these datasets were collected and linked to the human and mouse ENSEMBL genes (v.18). The final lists of human and mouse transcription factors contained 1,570 and 1,388 entries, respectively. The overlap comprised 1,334 entries.
For this project, we are selecting a subset of up to 200 human transcription factors for which phenotypes are known in mutant model organisms, but no target genes had been identified previously. Mainly, we are using reference data from mouse knock-out mutant databases, which have been already linked to orthologous human genes, for instance in the Genome Matrix interface (www.genomematrix.org). For the pilot knock down and transcriptome analysis experiments, we selected 3 transcription factors encoded by human chromosom 21 (BACH1: BTB and CNC homology 1, basic leucine zipper transcription factor 1; C21ORF66; PKNOX1: PBX/knotted 1 homeobox 1) for which the expression in the Hek293T17 cell line has been verified by qPCR.
2 - Design of RNAi molecules and knock down pilot experiments
When targeting endogenous gene expression, transfection efficiency is a critical parameter that may vary substantially depending on a combination of cell line and transfection reagent. Cationic liposome-based agents are commonly used for cellular transfection of oligonucleotides such as siRNA. We compared the transfection efficiency of Hek293T cells using Lipofectamine 2000 (Invitrogen) and the lipid formulation HiPerFect (Qiagen). With Lipofectamine 2000 we used the common transfection procedure of seeding cells 24h before the day of transfection. With HiPerFect we used the Fast-Forward protocol that allows cell seeding and transfection at the same day. In this second method cells are co-incubated with transfection complexes while in suspension before seeding into wells.
Using as reporter a scrambled control siRNA fluorescent labeled, we could measured the transfection efficiency and visualised the cellular distribution of the fluorescent molecules after transfection (Fig. 1). Using different siRNA and transfection reagent concentration, the transfection efficiency using HiPerFect and Fast-Forward protocol was always higher (85-90%) then with Lipofectamine 2000 and traditional protocol (75-80%). In both cases 48h post-transfection the siRNA accumulated in the cytoplasm near the nucleus.
We compared the silencing efficiency of siRNA (synthetic short interfering RNA) vs. esiRNA (endoribonuclease-prepared siRNA). siRNAs where purchased from Qiagen (2 different HP GenomeWide siRNA against each target gene) and from Invitrogen (3 different Stealth siRNA duplex oligoribonucleotides against each target gene). We designed and produced one dsRNA and RNaseIII digested esiRNA for each of the 3 pilot gene. Large (500-700 bp) dsRNA corresponding to the target region has been generated by in vitro transcription followed by RNase III digestion into short double strand RNA molecules.
We performed a series of transfection optimizations in Hek293T17 cell line for all silencing reagents and controls, at 48h post transfection. We evaluated the silencing performance experiments via qRT-PCR using Taq-Man approach.
We obtained the highest knock-down efficiencies when using HiPerFect as transfection reagent and when plating 1,3x105 cells/well together with the silencing complexes (Fig. 2).
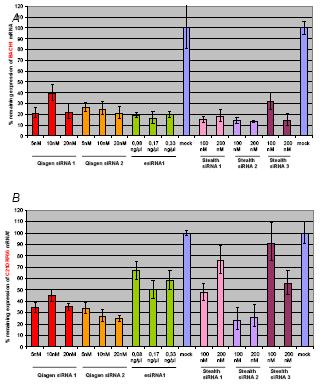
Fig 3: The indicated siRNAs and esiRNA were mixed with HiPerFect and added to Hek293T cells in suspension. Cells and silencing complexes were plated in 12 well plates. The final concentration of siRNA and esiRNA is shown. 48 hours after transfection, RNA from the treated cells was recovered and reverse transcribed. Target BACH1 (A) and C21ORF66 (B) cDNA levels were measured by real-time PCR using TaqMan assays. The expression of the target genes in the transfected cells was compared to cells transfected only with HiPerFect. Input cDNA in the different samples was normalized using real-time data for ß actin (ACTB). Error bars indicate SD.
Figure 3 shows the specific down-regulation of endogenous expression of BACH1 and C21ORF66 in Hek293T cells transfected with the three types of silencing molecules.
At least one of each kind of siRNA induced at least 75% knock-down of the target mRNA. The esiRNA against C21ORF66 mRNA designed and produced by us gave a maximum of 50% knock-down. We are currently generating a second esiRNA for each target gene using gene specific PCR products with T7 promoters purchased from RZPD (esiWay RNA Resource).
The silencing performances of the three different kind of RNAi molecules against the third pilot gene (PKNOX1) are still under analysis.
We are also now monitoring the protein knock-down by Western blot at 48h and 72h after transfection.
3 - Hybridisation and transcriptome analysis
Currently we are performing transcriptome analysis on human ENSEMBL cDNA macroarray using the RNA samples produced by knocking down the expression of BACH1 and C21ORF66 with the different RNAi silencing molecules.
Recent studies have indicated that transfection of cells with esiRNA and siRNA (at high concentration) can result in off-target effects, in which the RNAi molecules affect the expression of partially homologous gene targets. Moreover they can mediate a non specific interferon response. The macroarray results allow monitoring the toxicity of the silencing agents since the cDNAs corresponding to the main genes involved in the interferon response are spotted on the high-density nylon filters.
By performing the transcriptome analysis we will be able to check the specificity of each different silencing molecule (esiRNA, siRNA and Stealth siRNA) by testing at least two different siRNA and esiRNA against each target gene.
Lit.: All the future transfections of Hek293T cells will be done using HiPerFect transfection. For the next 15 target transcription factors (planned to be done by the end of this year) we will probably still use in parallel esiRNA and one kind of siRNA and we will perform the transcriptome analysis using the Ensemble cDNA chip and the Affymetrix platform. The knock-down performances together with the expression profiling data will enable us to evaluate which is be the most suitable RNAi molecule for our project to use to scale up the analysis for all the 150 transcription factors selected. The expression profiling data generated for the first 15 target transcription factors will already contribute significantly to the decipherment of the mechanisms regulating gene expression.


